8 Proteinases and Matrix Degradation
Extracellular Matrix Degrading proteinases
Aspartic Proteinases
Most aspartic proteinases have two aspartic acid residues in their catalytic sites, where the nucleophile that attacks the scissile peptide bond is an activated water molecule. Mammalian aspartic proteinases include the digestive enzymes (pepsin and chymosin), the intracellular cathepsin D and cathepsin E, and rennin. Among the proteinases belonging to this group, cathepsin D is the major aspartic proteinase involved in ECM degradation (Table 8-1). It exhibits proteolytic activity against most substrates such as aggrecan and collagen telopeptides with pH optima between pH 3.5 and 5. Because of the acidic pH optima and intracellular localization within lysosomes, cathepsin D is probably responsible for intracellular degradation of phagocytosed ECM fragments that previously were degraded in the extracellular spaces. A study on cartilage explant cultures using the aspartic proteinase inhibitor suggests, however, the possibility that cathepsin D secreted extracellularly contributes to the degradation of aggrecan in articular cartilage.1
Table 8-1 Proteinases That May Be Involved in Degradation of Extracellular Matrix
| Enzyme | Source | Inhibitor |
|---|---|---|
| Aspartic Proteinases | ||
| Cathepsin D | Lysosome | Pepstatin |
| Cysteine Proteinases | ||
| Cathepsin B | Lysosome | Cystatins |
| Cathepsin L | Lysosome | Cystatins |
| Cathepsin S | Lysosome | Cystatins |
| Cathepsin K | Lysosome | Cystatins |
| Calpain | Cytosol | Calpastatin |
| Serine Proteinases | ||
| Neutrophil elastase | Neutrophils | α1-PI |
| Cathepsin G | Neutrophils | α1-Antichymotrypsin |
| Proteinase 3 | Neutrophils | α1-PI, elafin |
| Plasmin | Plasma | Aprotinin |
| Plasma kallikrein | Plasma | Aprotinin |
| Tissue kallikrein | Glandular tissues | Aprotinin; kallistatin |
| tPA | Endothelial cells; chondrocytes | PAI-1; PAI-2 |
| uPA | Fibroblasts; chondrocytes | PAI-1; PAI-2; PN-1 |
| Tryptase | Mast cells | Trypstatin |
| Chymase | Mast cells | α1-PI |
| Metalloproteinases* | ||
| MMPs | Tissue cells; inflammatory cells | TIMP-1, 2, 3, and 4; RECK for MMP-2, 7, 9, and 14 |
| ADAMTSs | Tissue cells | TIMP-3 |
| ADAMs | Tissue cells; inflammatory cells | TIMP-3; RECK for ADAM10 |
ADAMs, a disintegrin and metalloproteinases; ADAMTSs, a disintegrin and metalloproteinases with thrombospondin motifs; MMPs, matrix metalloproteinases; PAI, plasminogen activator inhibitor; PI, proteinase inhibitor, PN, proteinase nexin; RECK, reversion-inducing, cysteine-rich protein with Kazal motifs; TIMP, tissue inhibitor of metalloproteinases; tPA, tissue-type plasminogen activator; uPA, urokinase-type plasminogen activator.
* For details of MMPs, ADAMTSs, and ADAMs, see Tables 8-2, 8-3, and 8-4.
Cysteine Proteinases
Cysteine proteinases are endopeptidases in which the nucleophile of the catalytic site is the sulfhydryl group of a cysteine residue. The ECM-degrading cysteine proteinases include lysosomal cathepsins B, L, S, and K and the calpains (see Table 8-1). Cathepsins B and L digest the telopeptide regions of fibrillar collagen types I and II, the nonhelical regions of collagen types IX and XI, and aggrecan at acidic pH. Cathepsin S has a similar spectrum of substrates within a broad range of pH values. Cathepsin K, also called cathepsin O, O2, or X, is a collagenolytic cathepsin that efficiently cleaves type I collagen at the triple helical regions at pH values between 4.5 and 6.6.2 The proteinase also degrades gelatin and osteonectin. On the basis of the data that cathepsin K is highly expressed by human osteoclasts3 and inactivating mutations or deletion of the gene result in an osteopetrotic phenotype in humans and animals, cathepsin K is believed to play a key role in bone resorption in joint diseases (see later). Because cathepsins B, L, S, and K are expressed in synovium or articular cartilage or both in rheumatoid arthritis and osteoarthritis, they may also be involved in the cartilage destruction through degradation of the ECM macromolecules.4
Calpains are Ca2+-dependent, papain-like cysteine proteinases and are ubiquitously distributed among mammalian cells. The best-characterized members of the calpain superfamily are µ-calpain and m-calpain, which also are called conventional (µ-calpain) and classic (m-calpain) calpains.5 Calpains are involved in various pathologic conditions such as muscle dystrophy by acting intracellularly. They are present in the extracellular spaces and in osteoarthritic synovial fluid, and they can degrade aggrecan.
Serine Proteinases
Serine proteinases require the hydroxyl group of a serine residue acting as the nucleophile that attacks the peptide bond. They include the largest number of proteinases classified to about 40 families. Most can degrade ECM macromolecules. The major ECM-degrading serine proteinases in joint tissues are subsequently described (see Table 8-1).
Neutrophil Elastase and Cathepsin G
Neutrophil elastase and cathepsin G cleave elastin; the telopeptide region of fibrillar collagen types I, II, and III; other collagen types IV, VI, VIII, IX, X, and XI; and other ECM components such as fibronectin, laminin, and aggrecan at neutral pH. These serine proteinases also can be involved indirectly in the breakdown of ECM by activating the zymogen of matrix metalloproteinases (proMMPs)6 and by inactivating endogenous proteinase inhibitors such as α2-antiplasmin, α1-antichymotrypsin, and tissue inhibitors of metalloproteinases (TIMPs).
Mast Cell Chymase and Tryptase
Chymase and tryptase are packaged in secretory granules together with histamine and other mediators in mast cells, which are infiltrated in rheumatoid synovium. Chymase is a chymotrypsin-like proteinase with a broad spectrum of activity against ECM components such as type VI collagen7 and aggrecan. It also activates proMMPs such as proMMP-1, proMMP-3, and proMMP-9.6 Although prochymase is activated intracellularly and stored in the granules, the activity in the granules is limited at low pH and becomes fully active when released extracellularly. Tryptase is a trypsin-like proteinase that degrades collagen type VI7 and fibronectin; it also activates proMMP-3.6
Plasmin and Plasminogen Activators
Plasminogen is synthesized in liver and secreted to plasma. It can bind to fibrin and to cells, and after activation by plasminogen activators, plasmin readily digests fibrin. Membrane-bound plasmin also degrades many ECM components including proteoglycan, fibronectin, type IV collagen, and laminin.8 Other important functions of plasmin are to initiate the activation of proMMPs,6 activate latent cell-associated transforming growth factor (TGF)-β1, and act as proenzyme convertase. Plasmin is generated through activation of plasminogen mainly by plasminogen activators, principally two serine proteinases, tissue-type plasminogen activator (tPA) and urokinase-type plasminogen activator (uPA).
The tPA is synthesized as a proenzyme of 70 kD and is secreted into the circulating blood primarily by endothelial cells, fibroblasts, chondrocytes, and tumor cells.8 In addition to being a major activator of plasminogen for fibrinolysis, tPA plays a key role in the clearance of fibrin from the circulation.
The uPA molecule was first purified from urine as a proenzyme of 54 kD.8 It is converted to the active form of two chains of 30 kD and 24 kD linked by a disulfide bond. Another fully active form of 33 kD is generated by plasmin. Although the expression of uPA is limited to certain cells such as renal tubules and bladder urothelium under physiologic conditions, it is more widely expressed in various cells including invasive cancer cells, migrating keratinocytes, and activated leukocytes in pathologic situations. Pro-uPA and two-chain uPA bind to a specific uPA receptor, a single-chain glycoprotein with a glycosylphosphatidylinositol (GPI) moiety expressed on fibroblasts, macrophages, and tumor cells. Receptor-bound uPA preferentially activates cell membrane-bound plasminogen into plasmin. Cell membrane-bound plasmin can activate receptor-bound pro-uPA. Among its specificities, uPA has a limited action on fibronectin.
Kallikreins
Two types of kallikreins, plasma and tissue kallikreins, are known. Plasma kallikrein, with two disulfide-linked chains (36 kD and 52 kD), is generated from prokallikrein of 88 kD by coagulation factor XIIa or by kallikrein itself. It activates kininogens to bradykinin and activates proMMP-1 and proMMP-3.6 Tissue kallikrein is synthesized in glandular tissues. It releases Lys-bradykinin from kininogen and activates proMMP-8.6
Metalloproteinases
Similar to aspartic proteinases, metalloproteinases are endopeptidases in which the nucleophilic attack on a peptide bond is mediated by a water molecule. A divalent metal cation, usually zinc, activates the water molecule. Among the metalloproteinases, MMPs (matrix metalloproteinases), which are also designated matrixins (a subfamily of the metzincin superfamily), are key ECM-degrading, zinc-dependent endopeptidases (Table 8-2). Accumulated evidence indicates, however, that some members of the ADAMTS (a disintegrin and metalloproteinase with thrombospondin motifs) family, which is an MMP-related gene family, also are involved in the degradation of ECM, especially cartilage proteoglycan (Table 8-3). Only a few members of the ADAM (a disintegrin and metalloproteinase) family have limited activity to ECM components (Table 8-4).
Table 8-2 Substrates of Human Matrix Metalloproteinases
| Enzymes | ECM Substrates | Non-ECM Substrates |
|---|---|---|
| Secreted-Type MMPs | ||
| Collagenases | ||
| MMP-1 (Interstitial collagenase) | Collagens I, II, III, VII, X; gelatins; aggrecan; link protein; entactin; tenascin; perlecan | α2-Macroglobulin; α1-PI; α1-antichymotrypsin; IGF-BP-2, -3, -5; pro-IL-1β; CTGF |
| MMP-8 (Neutrophil collagenase) | Collagens I, II, and III; gelatins; aggrecan; link protein | α1-PI |
| MMP-13 (Collagenase-3) | Collagens I, II, III, IV, IX, X, XIV; aggrecan; Fn; tenascin | CTGF; pro-TGF-β; α1-antichymotrypsin |
| Gelatinases | ||
| MMP-2 (Gelatinase A) | Gelatins; collagens IV, V, VII, XI; Ln; Fn; elastin; aggrecan; link protein | Pro-TGF-β; FGF receptor I; MCP-3; IGF-BP-5; pro-IL-1β; galectin-3; plasminogen |
| MMP-9 (Gelatinase B) | Gelatins; collagens III, IV, V; aggrecan; elastin; entactin; link protein | Pro-TGF-β; IL-2 receptor α; Kit-L; IGF-BP-3; pro-IL-1β; α1-PI; galectin-3; ICAM-1; plasminogen |
| Stromelysins | ||
| MMP-3 (Stromelysin-1) | Aggrecan; decorin; gelatins; Fn; Ln; collagens III, IV, IX, X; tenascin; link protein; perlecan | IGF-BP-3; pro-IL-1β; HB-EGF; CTGF; E-cadherin; α1-antichymotrypsin; α1-PI; α2-macroglobulin; plasminogen; uPA; proMMP-1, 7, 8, 9, 13 |
| MMP-10 (Stromelysin-2) | Aggrecan; Fn; Ln; collagens III, IV, V; link protein | ProMMP-1, 8, 10 |
| Matrilysins | ||
| MMP-7 (Matrilysin-1) | Aggrecan; gelatins; Fn; Ln; elastin; entactin; collagen IV; tenascin; link protein | Pro-α-defensin; Fas-L; β4 integrin; E-cadherin; pro-TNF; CTGF; HB-EGF; RANKL; IGF-BP-3; plasminogen |
| MMP-26 (Matrilysin-2) | Gelatin; collagen IV; Fn; fibrinogen | α1-PI; proMMP-9 |
| Furin-Activated MMPs | ||
| MMP-11 (Stromelysin-3) | Fn; Ln; aggrecan; gelatins | α1-PI; α2-macroglobulin; IGF-BP-1 |
| MMP-28 (Epilysin) | Unknown | Casein |
| Other Secreted-Type MMPs | ||
| MMP-12 (Metalloelastase) | Elastin; Fn; collagen V; osteonectin | Plasminogen; apolipoprotein A |
| MMP-19 (RASI-1) | Collagen IV; gelatin; Fn; tenascin; aggrecan; COMP; Ln; nidogen | IGF-BP-3 |
| MMP-20 (Enamelysin) | Amelogenin; aggrecan; gelatin; COMP | Unknown |
| MMP-21 | Unknown | Unknown |
| MMP-27 | Unknown | Unknown |
| Membrane-Anchored MMPs | ||
| Type I Transmembrane-Type MMPs | ||
| MMP-14 (MT1-MMP) | Collagens I, II, III; gelatins; aggrecan; Fn; Ln; fibrin; Ln-5 | ProMMP-2, 13; CD44; tissue transglutaminase |
| MMP-15 (MT2-MMP) | Fn; tenascin; nidogen; aggrecan; perlecan; Ln | ProMMP-2; tissue transglutaminase |
| MMP-16 (MT3-MMP) | Collagen III; Fn; gelatin | ProMMP-2; tissue transglutaminase |
| MMP-24 (MT5-MMP) | PG | ProMMP-2 |
| GPI-Linked MMPs | ||
| MMP-17 (MT4-MMP) | Gelatin; fibrinogen | Unknown |
| MMP-25 (MT6-MMP) | Gelatin; collagen IV; fibrin; Fn; Ln | ProMMP-2 |
| Type II Transmembrane-Type MMP | ||
| MMP-23 | Gelatin | Unknown |
COMP, cartilage oligomeric matrix protein; CTGF, connective tissue growth factor; ECM, extracellular matrix; Fn, fibronectin; GPI, glycosylphosphatidylinositol; HB-EGF, heparin-binding epidermal growth factor; IGF-BP, insulin-like growth factor binding protein; ICAM-1, intercellular adhesion molecule 1; RANKL, receptor activator for nuclear factor κB ligand; IL-1, interleukin-1; Ln, laminin; MCP, monocyte chemoattractant protein; PG, proteoglycan; PI, proteinase inhibitor; TGF, transforming growth factor; TNF, tumor necrosis factor; uPA, urokinase-type plasminogen activator.
Matrix Metalloproteinases
The human MMP family comprises 23 different members, which have MMP designations (numbered according to a sequential numbering system) and common names coined by the authors of the published reports (see Table 8-2). On the basis of the biochemical properties provided by the domain structures and on their substrate specificity, these family members are classified into two major subgroups: secreted-type MMPs and membrane-anchored MMPs. MMP-4, MMP-5, and MMP-6 are excluded from the list because they are identical to other known MMPs (i.e., MMP-3 and MMP-2). MMP-18 and MMP-22 also are missing in Table 8-2 because they are assigned to Xenopus collagenase-4 and chicken MMP.
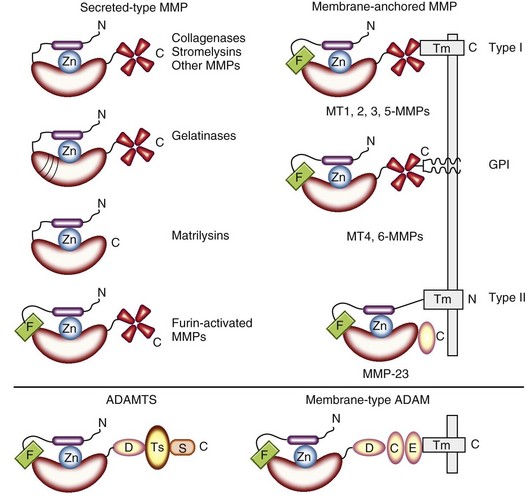
Most secreted-type MMPs including collagenases, stromelysins, and other MMPs are composed of three basic domains—the propeptide, catalytic, and hemopexin-like domains—that are preceded by hydrophobic signal peptides (Figure 8-1). The N-terminal propeptide domain has one unpaired cysteine in the conserved sequence of PRCGXPD. The cysteine residue in the sequence interacts with the essential zinc atom in the catalytic domain to prevent it from binding the catalytic water molecule, maintaining the proenzyme in an inactive state. The catalytic domain has the zinc-binding motif HEXGHXXGXXH, in which three histidines bind to the catalytic zinc atom. The four-blade, C-terminal hemopexin-like domain, which is connected to the catalytic domain by a proline-rich hinge region, interacts with ECM components and can play a role in determining the substrate specificity in some MMPs. Gelatinases have these domains with additional insertions of collagen-binding type II repeats of fibronectin in the catalytic domain (see Figure 8-1); this provides them with collagen-binding properties. Matrilysins are the shortest, lacking the hemopexin-like domains. Furin-activated MMPs contain insertions of a basic motif with a cleavage site by proprotein convertases including furin at the end of the propeptide domains (see Figure 8-1).
Membrane-anchored MMPs include three different types of MMPs: type I transmembrane-type MMPs, GPI-linked MMPs, and type II transmembrane-type MMP. MMP-14, MMP-15, MMP-16, and MMP-24 (MT1-, MT2-, MT3-, and MT5-MMPs, respectively) have type I transmembrane domains in the C-terminal region, but MMP-17 (MT4-MMP) and MMP-25 (MT6-MMP) contain GPI anchors in the C-terminal region without transmembrane domains (see Figure 8-1). MMP-23 is unique in that it has type II transmembrane domain, a cysteine array, and an immunoglobulin-like domain instead of a hemopexin-like domain (see Figure 8-1).
Secreted-Type Matrix Metalloproteinases
Collagenases (MMP-1, MMP-8, and MMP-13)
The collagenases include MMP-1 (interstitial collagenase, collagenase-1), MMP-8 (neutrophil collagenase, collagenase-2), and MMP-13 (collagenase-3). These MMPs attack triple helical regions of interstitial collagen types I, II, and III at a specific single site after the Gly residue of the partial sequences Gly-(Ile or Leu)-(Ala or Leu), located about three-fourths of the distance from the N-terminus. This cleavage generates fragments approximately three-fourths and one-fourth of the size of the collagen molecules. A biochemical study has disclosed the molecular mechanism of the cleavage: MMP-1 unwinds the triple helical structure by interacting with the α2(I) chain of type I collagen and cleaves the three α chains in succession.9 MMP-13 is unique in that it cleaves α chains of type II collagen at two sites of the Gly906-Leu907 Gly909-Gln910 bonds.10 All of these collagenases degrade the interstitial collagens, but their specific activities against the collagens are different; MMP-1, MMP-8, and MMP-13 preferentially digest collagen types III, I, and II.10,11 Although rodents such as mice were originally thought to have only two collagenases (MMP-8 and MMP-13) and to lack the MMP-1 gene, rodent homologues of the human MMP-1 gene were cloned and named mouse collagenase A and B (Mcol-A and Mcol-B).12
In addition to the interstitial fibrillar collagens, MMP-1, MMP-8, and MMP-13 degrade many other ECM macromolecules. MMP-1 digests entactin, collagen X, gelatins, perlecan, aggrecan, and cartilage link protein (see Table 8-2). MMP-8 digests aggrecan, gelatins, and cartilage link protein (see Table 8-2). MMP-13 hydrolyzes aggrecan; types IV, IX, X, and XIV collagens; fibronectin; and tenascin. Non-ECM substrates of MMP-1, MMP-8, and MMP-13 include α2-macroglobulin, α1-antiproteinase inhibitor, α1-antichymotrypsin, insulin-like growth factor binding protein (IGF-BP)-2 and IGF-BP-3, connective tissue growth factor (CTGF), and pro-TGF-β (see Table 8-2).
Gelatinases (MMP-2 and MMP-9)
MMP-2 (gelatinase A) and MMP-9 (gelatinase B) belong to the gelatinase subgroup. Both MMPs readily digest gelatins and cleave collagen types IV and V.13,14 Elastin, aggrecan, and cartilage link protein also are substrates of the gelatinases. Although MMP-2 and MMP-9 share such substrates, they have different activities on several ECM macromolecules. MMP-2, but not MMP-9, digests fibronectin and laminin,13 and type III collagen and α2 chains of type I collagen are degraded only by MMP-9.14 The gelatinases also process directly TGF-β into an active ligand (see Table 8-2). MMP-2 and MMP-9 cleave fibroblast growth factor receptor type I and interleukin (IL)-2 receptor type α (see Table 8-2). MMP-9 also releases soluble Kit-ligand.15 MMP-2 processes monocyte chemoattractant protein (MCP)-3 into an MCP-3 fragment deleting the N-terminal four amino acids, which can bind to CC-chemokine receptors and act as a general chemokine antagonist.16
Stromelysins (MMP-3 and MMP-10)
The subgroup of stromelysins consists of MMP-3 (stromelysin-1) and MMP-10 (stromelysin-2). They share 78% identity in amino acid sequence and have similar enzymatic properties.17 The enzymes hydrolyze numerous ECM macromolecules including aggrecan, fibronectin, laminin, and collagen IV (see Table 8-2).18 Collagen types III, IX, and X and telopeptides of collagen types I, II, and XI also are digested by MMP-3.19 In addition to the ECM components, MMP-3 is active on IGF-BP-3, IL-1β, heparin-binding epidermal growth factor (HB-EGF), CTGF, E-cadherin, α1-antichymotrypsin, and α1-proteinase inhibitor (see Table 8-2). MMP-3 also activates many proMMPs.6 A similar activator function has been identified for MMP-10.20
Matrilysins (MMP-7 and MMP-26)
Matrilysins include MMP-7 (matrilysin-1) and MMP-26 (matrilysin-2), which are the smallest of the MMPs, having only the propeptide and catalytic domains. The substrate specificity of MMP-7 is similar to that of stromelysins, digesting numerous ECM components including aggrecan; gelatins; fibronectin; laminin; elastin; entactin; collagen types III, IV, V, IX, X, and XI; fibrin/fibrinogen; vitronectin; tenascin; and link protein (see Table 8-2). Although these substrates overlap with the substrates of other MMPs, the specific activity of MMP-7 to most substrates is highest among the MMPs.21,22 Non-ECM molecules such as α-defensin, Fas ligand, β4 integrin, E-cadherin, plasminogen, tumor necrosis factor (TNF), and CTGF also are the substrates for MMP-7 (see Table 8-2). MMP-26 degrades gelatin, type IV collagen, fibronectin, fibrinogen, and α1-proteinase inhibitor,23–25 but information about other substrates is still limited.
Furin-Activated Matrix Metalloproteinases (MMP-11 and MMP-28)
MMP-11 (stromelysin-3) and MMP-28 (epilysin) contain an RKRR sequence at the end of the propeptide, which is a unique motif for intracellular processing of proproteins to mature molecules by furin and other proprotein convertases. ProMMP-11 is activated intracellularly by the action of furin.26 MMP-11 shows only weak proteolytic activity against gelatin, laminin, fibronectin, and aggrecan,27 but it has respectable catalytic action in digesting α1-proteinase inhibitor, α2-macroglobulin, and IGF-BP-1 (see Table 8-2).28,29 MMP-28 can degrade casein, but its natural substrates are unknown.30
Other Secreted-Type Matrix Metalloproteinases (MMP-12, MMP-19, MMP-20, MMP-21, and MMP-27)
MMP-12 (metalloelastase),31 MMP-19 (RASI-1),32 MMP-20 (enamelysin),33 MMP-21,25 and MMP-27 have structural characteristics similar to collagenases and stromelysins. These MMPs are not classified into the previously mentioned subgroups, however, because their substrates and other biochemical characters are not fully examined at present. Overall information on the substrate specificity of MMP-12,31 MMP-19,34,35 and MMP-2033,35 suggests that they are stromelysin-like proteinases. MMP-1231 digests elastin, fibronectin, collagen V, osteonectin, and plasminogen (see Table 8-2). MMP-19, which was originally reported as MMP-18 but renamed as MMP-19, cleaves type IV collagen, laminin, fibronectin, gelatin, tenascin, entactin, fibrin/fibrinogen, aggrecan, and cartilage oligomeric matrix protein (COMP) (see Table 8-2).34,35 MMP-20 also digests amelogenin, aggrecan, and COMP.35 Substrates of MMP-21 and MMP-27 are unknown, however.
Membrane-Anchored Matrix Metalloproteinases
Type I Transmembrane-Type MMPs
These include MMP-14 (MT1-MMP),36 MMP-15 (MT2-MMP),37 MMP-16 (MT3-MMP),38 and MMP-24 (MT5-MMP).39 All of these MT-MMPs can activate proMMP-2, but MMP-14 may play a major role in the activation of proMMP-2 in various tissues (see later). Besides the activator function, however, MMP-14 digests the triple helical portions of interstitial collagen types I, II, and III and other ECM components including fibronectin, laminin, aggrecan, and gelatin (see Table 8-2).40 MMP-15 also digests fibronectin, tenascin, nidogen, aggrecan, perlecan, and laminin.41 MMP-16 cleaves collagen type III, fibronectin, and gelatins.42 MMP-17 (MT4-MMP)43 and MMP-25 (MT6-MMP)44 are GPI-linked MMPs. MMP-17 and MMP-25 can digest gelatin and fibrin/fibrinogen (see Table 8-2).44–46 MMP-23 (cysteine array-MMP, MIFR) is a type II transmembrane-type MMP47; almost identical genes are cloned, called MMP-23A and MMP-23B. MMP-23 digests gelatin,48 but no information about other substrates is available (see Table 8-2). A unique aspect of MMP-23 is that this MMP is expressed in only female and male reproductive organs such as endometrium, ovary, testis, and prostate,48 but its functions are not well established.
ADAM and ADAMTS Families
Two ADAM (a disintegrin and metalloproteinase) gene families exist: membrane-type ADAM with transmembrane domain (ADAM) and secreted-type ADAM with thrombospondin motifs (ADAMTS) (see Figure 8-1). The active sites in the catalytic domains of most members of both gene families contain a common sequence of HEXGHXXGXXHD with the “Met-turn,” which also is present in MMP members.
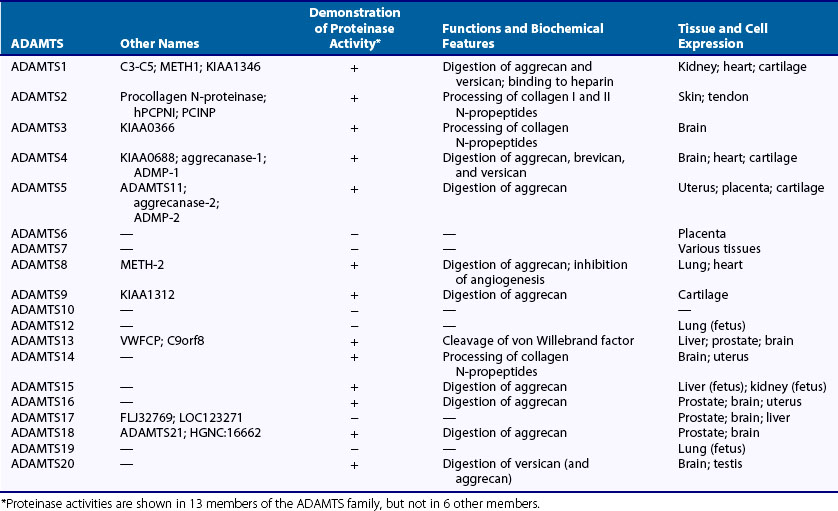
The ADAMTS family includes 19 members. Although information about substrates and biologic functions is still limited, ADAMTS1, ADAMTS2, ADAMTS3, ADAMTS4, ADAMTS5, ADAMTS8, ADAMTS9, ADAMTS14, ADAMTS15, ADAMTS16, ADAMTS18, and ADAMTS20 all are ECM-degrading proteinases (see Table 8-3). ADAMTS1,49 ADAMTS4,50 ADAMTS5,51 ADAMTS8, ADAMTS9, and ADAMTS15 can preferentially cleave aggrecan at the five Glu-X bonds including the Glu373-Ala374 bond (the aggrecanase site). Because of aggrecan-degrading activity, ADAMTS4 and ADAMTS5 are named aggrecanase-1 and aggrecanase-250,51; versican also is digested by these proteinases,52 and brevican is cleaved by ADAMTS4 (see Table 8-3).53 The C-terminus-truncated ADAMTS4 also degrades fibromodulin and decorin.54 ADAMTS16, ADAMTS18, and ADAMTS20 also appear to have weak aggrecanase activity. ADAMTS2 and ADAMTS3 process the N-terminal propeptides of type I and II collagens and are named procollagen N-proteinase. Activity of procollagen N-proteinase also is known with ADAMTS14. ADAMTS13 is a von Willebrand factor–cleaving proteinase, and its mutation causes thrombotic thrombocytopenic purpura. Proteinase activities of other ADAMTS species are still unknown.
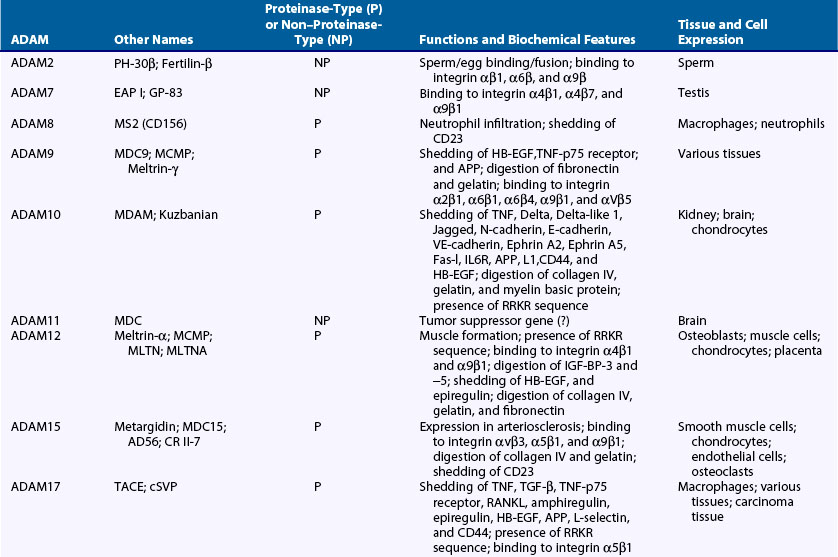
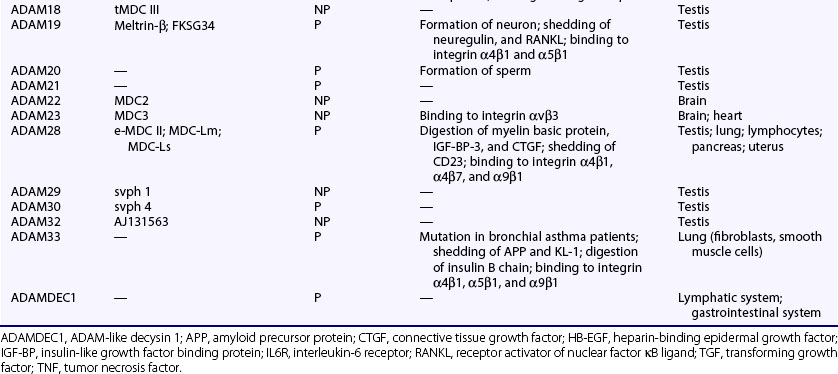
The human genome contains 25 ADAM genes including 4 pseudogenes, and thus the human ADAM family is composed of 21 members (Table 8-4). Among the ADAMs, ADAM8, ADAM9, ADAM10, ADAM12, ADAM15, ADAM17, ADAM19, ADAM20, ADAM21, ADAM28, ADAM30, ADAM33, and ADAMDEC1 exhibit proteolytic activity (proteinase-type ADAMs) (see Table 8-4). Although ADAM10, ADAM12, and ADAM15 degrade type IV collagen, the main substrates of these ADAMs are various membrane proteins, which include precursors of cytokines and growth factors such as TNF, HB-EGF and neuregulin, IGF-BPs, receptors such as p75 TNF receptor, IL-1 receptor II, and other membrane proteins related to development such as Notch ligand and ephrin (see Table 8-4).55–60 According to these data, a major function of the ADAMs is shedding of the membrane proteins. ADAM17 cleaves a proform of TNF at the physiologic processing site into the soluble form of TNF and is called TNF-converting enzyme. ADAM17 also is involved in release of L-selectin, TGF-α, and p75 TNF receptor.55 ADAM9, ADAM12, and ADAM17 can shed HB-EGF from its precursor. ADAM12 and ADAM28 cleave IGF-BP-3 and IGF-BP-5.60,61 CD23 is shed by ADAM8, ADAM15, and ADAM28.62 Other functions of ADAMs include binding to integrins, cell-cell interaction, cell migration, and signal transduction (see Table 8-4).63
Stay updated, free articles. Join our Telegram channel

Full access? Get Clinical Tree