18 Innate Immunity
Innate immunity depends on recognition of conserved molecular patterns found in many microorganisms.
Antimicrobial peptides are important effectors of innate immunity.
Innate immune responses have a strong impact on the development of adaptive immunity.
It has become common practice in immunology to divide the mechanisms involved in host defense into adaptive and innate components; this provides a useful framework for classifying the numerous cells, receptors, and effector molecules that combine to make up the vertebrate immune system (Table 18-1). A specific immune response, such as the production of antibodies or T cells against a particular pathogen, is referred to as adaptive immunity because it represents an adaptation that occurs during the lifetime of an individual as a result of exposure to that pathogen. Adaptive immune responses involve the clonal expansion of T and B lymphocytes bearing a large repertoire of somatically generated receptors that can be selected to recognize virtually any pathogen. The adaptive immune system of any given individual is profoundly molded by the immunologic challenges encountered by that individual during the course of a lifetime. A hallmark of adaptive immune responses is that they are highly specific for the triggering agent, and they provide the basis for immunologic memory. This property of memory endows the adaptive immune response with its “anticipatory” property, which provides increased resistance against future infection with the same pathogen and also allows vaccination against future infectious threats.
Table 18-1 Contrasting Features of Innate and Adaptive Immune Systems
| Property | Innate Immune System | Adaptive Immune System |
|---|---|---|
| Receptors | Relatively few (several hundred?) | Many (potentially 1014 or more) |
| Fixed in genome | Encoded in gene segments | |
| Gene rearrangement not required | Gene rearrangement required | |
| Distribution | Nonclonal | Clonal |
| All cells of a class identical | All cells of a class distinct | |
| Targets | Conserved molecular patterns | Details of molecular structure |
| Lipopolysaccharides | Proteins | |
| Lipoteichoic acids | Peptides | |
| Glycans and peptidoglycans | Carbohydrates | |
| Others | ||
| Self–non–self-discrimination | Perfect: selected over evolutionary time | Imperfect: selected in individual somatic cells |
| Action time | Immediate or rapid (seconds to hours) | Delayed (days to weeks) |
| Response | Microbicidal effector molecules | Clonal expansion or anergy of specific T and B lymphocytes |
| Antimicrobial peptides | Cytokines (IL-2, IL-4, IFN-γ, others) | |
| Superoxide | Specific antibody production | |
| Nitric oxide | Specific cytolytic T cell generation | |
| Cytokines (IL-1, IL-6, others) | ||
| Chemokines (IL-8, others) |
IFN, interferon; IL, interleukin.
Modified from Medzhitov R, Janeway CA Jr: Innate immune recognition, Annu Rev Immunol 20:197, 2002.
Evolutionary Origins of Innate Immunity
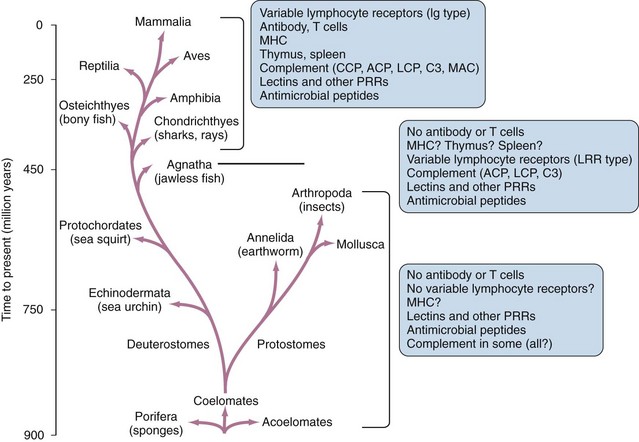
In spite of its obvious importance for most vertebrate organisms, the adaptive immune system is a relatively recent evolutionary development (Figure 18-1). In the great majority of present-day vertebrate species, the adaptive immune system is based on the ability to generate large families of variable lymphocyte receptors with immunoglobulin-like structures. This ability has been conserved owing to the acquisition of a specialized recombination system that mediates the assembly of gene segments in the T cell and B cell receptor families, which most likely occurred through invasion of the genome of a primitive vertebrate by a transposable element or virus carrying this machinery.1 This critical step in the evolution of the immune system can be traced back to the emergence of the ancestors of present-day jawed fish, which represent the most primitive extant species known to have adaptive immune systems based on the generation of large families of specific immunoglobulin-type receptors.2 Recently, other systems of variable lymphocyte receptors that are unrelated to immunoglobulins but also provide the basis for an adaptive immune response have been discovered in primitive jawless fish such as lampreys and hagfish.3 This finding shows that at least two different strategies for the creation of an adaptive immune system emerged at the dawn of vertebrate evolution about 500 million years ago, and it emphasizes the importance of adaptive immunity for survival and further evolution of the vertebrate lineages.
Given this key role of adaptive immunity in the evolution and survival of vertebrates, it is surprising that all invertebrate animals, and possibly some of the lowest vertebrate species as well, completely lack the ability to generate lymphocyte populations bearing large families of clonally diverse antigen receptors.4,5 In these animals, protection against pathogen invasion depends entirely on innate immunity, elements of which appear to exist in all animals and plants and must have evolved with the earliest multicellular forms of life. In many cases, components of the innate immune system are significantly conserved in structure and function in animals from the lowliest invertebrates to the most complex vertebrates.4 This preservation of innate immune mechanisms, with their functions largely intact, over such vast evolutionary distances is a clear indication of their importance, even in animals that have developed sophisticated adaptive immune responses.
PAMPs and DAMPs: Patterns for Innate Immune Recognition
Pathogen-Associated Molecular Patterns
In addition to allowing direct recognition of molecules produced by various microorganisms, the innate immune system is able to respond to the patterns of host-derived molecules released by cells undergoing necrotic death. The molecules recognized are generally referred to as damage-associated molecular patterns (DAMPs), and include multiple different families of proteins, as well as nonproteinaceous substances such as uric acid microcrystals.6,7 Thus, the response to DAMPs can be an indirect response to microbial invasion, or it can be triggered by other types of tissue damage such as ischemia to result in sterile inflammation.
Pattern Recognition Receptors
Recognition of PAMPs and DAMPs is mediated by a collection of germline-encoded molecules known collectively as pattern recognition receptors (PRRs) (Table 18-2). These receptors are host proteins that have evolved, through many millions of years of natural selection, to possess precisely defined specificities for particular PAMPs or DAMPs expressed by microorganisms. The total number of PRRs present in complex vertebrates such as humans is estimated to be several hundred—a number limited by the size of the genome of any animal and the number of genes it can dedicate to immune protection. The human genome, for example, is estimated to contain approximately 20,000 to 35,000 genes, most of which are not related directly to the immune system. This demonstrates one of the strong points of contrast between innate and adaptive immune systems, because the latter can possess in the range of 1014 different somatically generated receptors for foreign antigens in the form of antibodies and T cell receptors. With its much more limited array of receptors, the innate immune system uses the strategy of targeting highly conserved PAMPs that are shared broadly by large classes of microorganisms. Because most pathogens contain PAMPs, this strategy allows the generation of at least partial immunity against most infections.
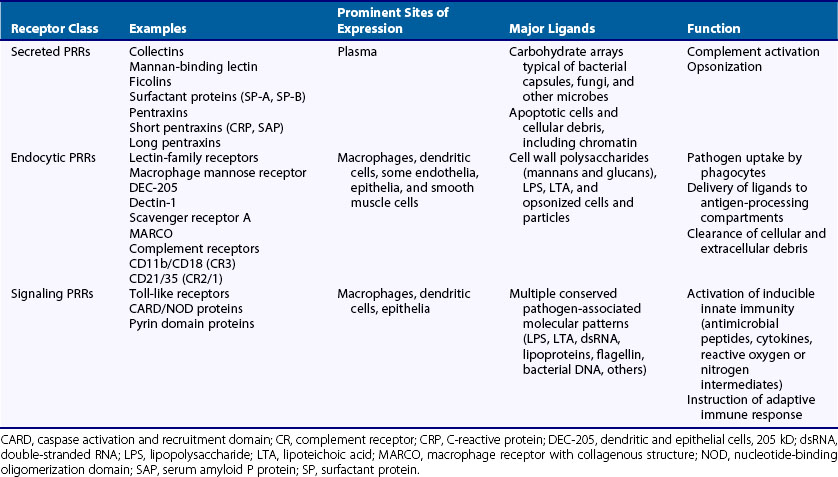
In recent years, considerable progress has been made toward identifying many of the important PRRs involved in the induction of innate immunity. These receptors can be classified into three functional classes: secreted, endocytic, and signaling PRRs (see Table 18-2). In addition, many of the known PRRs can be classified into structurally defined families on the basis of a few characteristic protein domains. Among these, the best known include proteins with calcium-dependent lectin domains, scavenger receptor domains, and leucine-rich repeat domains.
Pattern Recognition Receptors of the Lectin Family
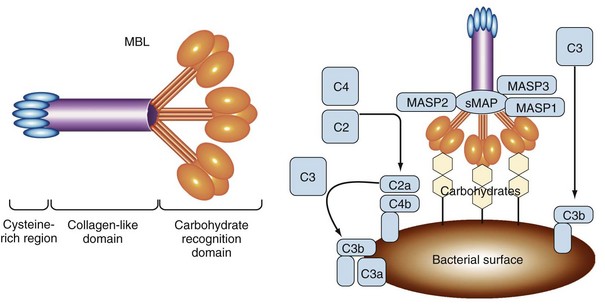
Calcium-dependent lectin domains are common modules of secreted and membrane-bound proteins involved in the binding of carbohydrate structures. A well-characterized PRR belonging to this class is the mannan-binding lectin (MBL), also known as soluble mannose-binding protein, which represents a secreted PRR that functions in initiation of the complement cascade (Figure 18-2).8,9 This protein is synthesized primarily in the liver on a constitutive basis, although its production can be increased as an acute phase reactant following many types of infection. MBL binds to carbohydrates on the outer membranes and capsules of many bacteria, as well as fungi, some viruses, and parasites. Although mannose and fucose sugars bound by MBL can also be found on the surfaces of normal mammalian cells, they are present at too low a density or in the wrong orientation to efficiently engage the lectin domains of MBL. In contrast, the coats of many microorganisms contain an array of these sugars, which allows strong binding of MBL. Thus, in this case, the spacing and orientation of specific carbohydrate residues constitute the PAMP that triggers the activation of innate immunity by MBL. MBL functions as one of a small number of secreted PRRs that can initiate the lectin pathway of complement activation. At least two other soluble proteins with lectin activity in human plasma, known as ficolins (ficolin/P35 and H-ficolin), can also activate this pathway following their interaction with bacterial polysaccharides.10
Several of the soluble lectin-type PRRs also play an important role in the opsonization of microbes by binding to their surfaces and directing them to receptors on phagocytic cells. Among these are two pulmonary surfactant proteins, SP-A and SP-D, which similarly recognize and bind to the surface sugar codes of microbes in the respiratory tract.11 These molecules are similar in structure to MBL, having both collagen-like and lectin domains, and together they constitute a family of soluble PRRs known as collectins. Another family of soluble PRRs that performs a similar function in plasma is the pentraxins, so called because they are formed by the association of five identical protein subunits.12 This family includes the acute phase reactants C-reactive protein (CRP) and serum amyloid P protein (SAP), along with a number of so-called long pentraxins, which have an extended polypeptide structure with homology to the classic short pentraxins (i.e., CRP and SAP) only at their carboxy-terminal domains. Long pentraxins are expressed in a variety of different tissues and cells, and their specific functions are mostly unknown. However, the long pentraxin PTX3 has been shown to play an important, nonredundant role in resistance to fungal infection in mice, and recent studies indicate that PTX3 is essentially a functional ancestor of antibodies that recognizes microbes and promotes their clearance through complement activation and phagocytosis.13
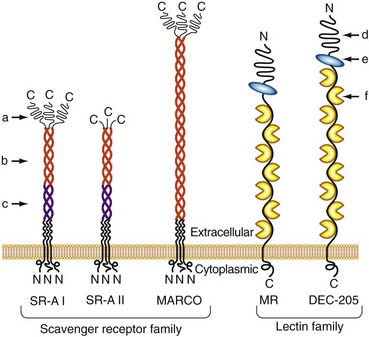
In addition to these soluble proteins, a large number of membrane-bound glycoproteins with lectin domains are known to exist; some of them participate in innate immunity by serving as endocytic PRRs for the uptake of microbes or microbial products14,15 (Figure 18-3). One of the most extensively studied of these is the macrophage mannose receptor (MMR).16 Although originally identified on alveolar macrophages and known to be expressed on macrophage subsets throughout the body, this receptor is also expressed on a variety of other cell types, including certain endothelia, epithelia, and smooth muscle cells. The MMR is a membrane-anchored, multilectin domain–containing protein that mediates the binding of a broad range of pathogens, leading to their internalization via endocytosis and phagocytosis. Although the major function of the MMR appears to be directing the uptake of its ligands, evidence suggests that this receptor may be capable of signaling to modify macrophage functions following receptor engagement.17 Another member of this receptor family, the β-glucan binding cell-surface lectin known as dectin-1, has a role in the modulation of inflammation in a mouse model of infection-induced arthritis.18
Pattern Recognition Receptors of the Scavenger Receptor Family
The scavenger receptor family contains a broad range of structurally diverse cell surface proteins that are expressed most prominently on macrophages, dendritic cells, and endothelial cells19 (see Figure 18-3). Although they were originally defined by their ability to bind and take up modified serum lipoproteins, they also bind a wide range of other ligands, including bacteria and some of their associated products. Multiple members of this family have been implicated as PRRs for innate immunity. These include the scavenger receptor A (SR-A) and a related molecule called the macrophage receptor with collagenous structure (MARCO).20 Both of these molecules contain a scavenger receptor cysteine-rich domain in the distal ends of their membranes and a collagen-like stalk with a triple-helical structure. Both are known to bind bacteria, and SR-A also binds well-known PAMPs such as lipoteichoic acids and LPS.21,22 Mice that have been made deficient in SR-A by targeted gene disruption show increased susceptibility to infections caused by a variety of bacteria, thus providing strong evidence of the role of scavenger receptors in protective immunity, most likely through the activation of innate immune mechanisms.23,24 Members of the class B scavenger receptor family, including CD36 and SR-BI/CLA-1, have also been found to recognize a variety of pathogen-derived molecules.19 Although these members of the scavenger receptor family clearly function as endocytic PRRs in the uptake of microbes, their potential to serve as signaling receptors has not yet been established. However, several scavenger receptors play a co-receptor role in the signal transduction process mediated by members of the Toll-like receptor (TLR) family (discussed later), most likely by capturing specific ligands and transferring them to adjacent TLRs.19
Pattern Recognition Receptors with Leucine-Rich Repeat Domains
Leucine-rich repeat domains (LRRs) are structural modules found in many proteins, including PRRs involved in signaling the activation of innate immunity. Molecules in this class include, most notably, the family of mammalian Toll-like receptors (TLRs), which are membrane-bound signal-transducing molecules that play a central role in the recognition of extracellular and vacuolar pathogens.25 Two families of cytoplasmic LRR-containing receptors have also been identified; they play a prominent role in the innate immune recognition of PAMPs expressed by intracellular pathogens. These include the families of caspase activation and recruitment domain (CARD) proteins and of pyrin domain proteins.26 Molecules are closely related in structure and function to proteins found in invertebrates and plants involved in pathogen resistance, highlighting the ancient origin of these pathways for host defense; they appear to have been recognizably conserved throughout approximately 1 billion years of evolution.
Toll-like Receptors
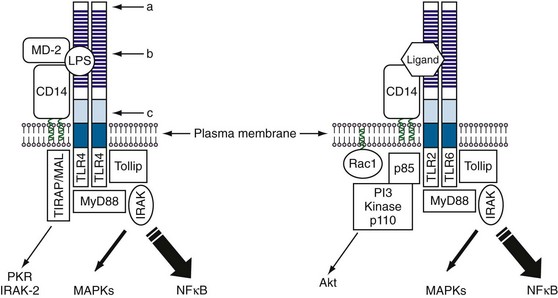
The first member of the Toll family to be discovered was the Drosophila Toll protein, which was identified as a component of a signaling pathway that controls dorsoventral polarity during development of the fly embryo.27 The sequence of Toll showed it to be a transmembrane protein with a large extracellular domain containing multiple tandemly repeated LRRs at the N-terminal end, followed by a cysteine-rich domain and an intracellular signaling domain (Figure 18-4). A role for Toll in immune responses was suggested by the observation that its intracellular domain shows homology to the mammalian interleukin-1 receptor (IL-1R) cytoplasmic domain.28 This association was later confirmed in studies showing that Toll was critical for the antifungal response in the fly, linking this pathway for the first time to innate immunity.29 Identification of Drosophila Toll eventually led to a search for similar proteins in mammals; this effort has been richly rewarded, yielding a family of 10 Toll-like receptors (TLRs) in humans and 12 in mice.30 Among these, TLR1 through TLR9 are conserved between mice and humans, TLR10 is present only in humans, and TLR11 through TLR13 are expressed only in mice.30 All these molecules contain large extracellular domains with multiple LRRs, as well as intracellular signaling domains known as Toll/IL-1R, or TIR, domains.31 Many of these TLRs have been linked to innate immune responses against various PAMPs of different microorganisms.32
< div class='tao-gold-member'>
Stay updated, free articles. Join our Telegram channel

Full access? Get Clinical Tree