Fig. 8.1
Distinct domain structures and differential cell localization of ADAMTS and ADAM proteinases. The domain structures of ADAMTS5 and of a typical ADAM proteinase are shown in the context of the cell surface, with the various domains indicated. Note the transmembrane insertion of ADAMs. ADAMTS proteinases are secreted, but several bind near the cell surface, potentially through components of pericellular matrix or to cell-surface molecules
Once genome sequencing of many organisms was completed, it became apparent that the repertoire of genes encoding for extracellular matrix had expanded substantially in vertebrates (Huxley-Jones et al. 2009), which intuitively suggests the reason for an observed concomitant expansion of genes encoding metzincins (Huxley-Jones et al. 2007). Furthermore, tissue inhibitors of metalloproteinases (TIMPs), the main endogenous inhibitors of metalloproteinases, evolved from a single gene in Drosophila into the four human TIMPs studied today (Brew and Nagase 2010), further substantiating the significance of proteolysis in advanced biologic systems.
MMPs have been extensively studied in orthopaedic biology (Pasternak and Aspenberg 2009). For the purpose of this chapter, we will focus only on ADAMTS metalloproteinases and summarize their emerging roles in the intervertebral disc. ADAMs are mentioned where relevant, since they are often wrongly thought to be the same as ADAMTS proteinases yet have been neglected in the context of the disc.
The first ADAMTS protease (ADAMTS1) was identified 15 years ago by Kuno et al. (1997), as an inflammation associated gene product and initially thought to be a variant ADAM since it had the reprolysin-type catalytic domain (Kuno et al. 1997). Subsequent molecular cloning of 18 structurally similar proteinases, facilitated greatly by rapid progress in the human genome project, demonstrated the existence of this hitherto unknown metalloproteinase family as one that is distinct from ADAMs. It should be noted that the enzyme designated as ADAMTS11 (Abbaszade et al. 1999) is now referred to as ADAMTS5, and the designation ADAMTS11 is left vacant. Thus, although 20 ADAMTS numbers are assigned, there are 19 ADAMTS proteinases.
8.2 ADAMTS Structure and Function
Once the full repertoire of ADAMTS proteinases was determined, evolutionary analysis showed that they clustered into several distinct subgroups (Apte 2004; Huxley-Jones et al. 2005) (Fig. 8.2). Typically, ADAMTS proteinases within these subgroups have similar domain structures, high sequence similarity, and sufficient functional overlap that they work cooperatively in certain contexts. The ADAMTS ancillary domain consists of a disintegrin-like module, a thrombospondin type 1 repeat (TSR), a cysteine-rich region, a cysteine-free spacer region, one or more additional TSRs, as well as other modules (Apte 2009) (Fig. 8.2). The differences in the ancillary domains allow each proteinase to bind and act on distinct substrates, and with very few exceptions, ADAMTS catalytic domains without adjoining ancillary domains lack proteolytic activity. The detailed structure and posttranslational modification of ADAMTS proteases were previously reviewed (Apte 2009). Figure 8.1 contrasts the domain structure of a typical ADAMTS protease (ADAMTS5) with a prototypic ADAM, illustrating also the different localization with respect to cells.
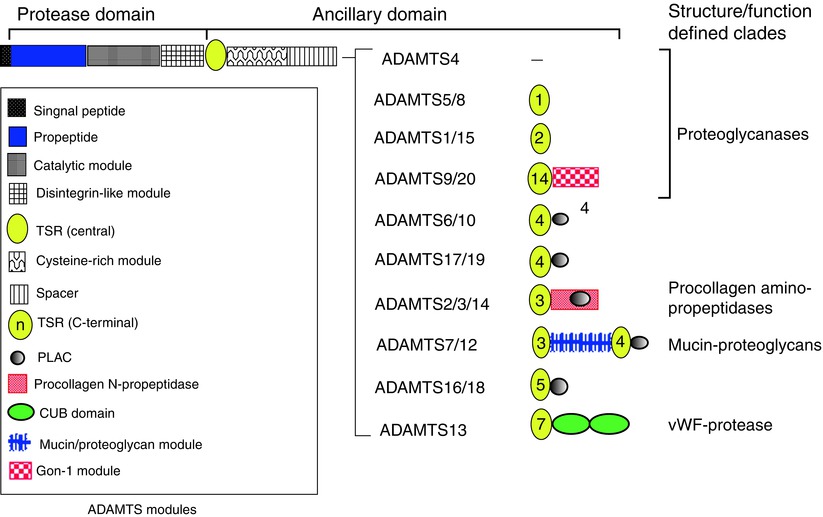

Fig. 8.2
Mammalian ADAMTS proteases. The domain backbone shared by each ADAMTS protease is shown at the top. The unique structure of each ADAMTS protease C-terminal to the backbone is indicated on the right, and the key to these modules is located at left. Some clades are named according to structural or functional characteristics that best define them; clades without a known function or a defining characteristic are not named. The proteoglycanases constitute a super-clade comprising ADAMTS proteases with different domain structures (The figure is based on reference sequences obtained from GenBank)
A high degree of regulation of ADAMTS proteinases is a crucial aspect of their biology. Proprotein convertases such as furin cleave ADAMTS and ADAM proteinases after paired basic residues at the junction of the propeptide and catalytic domains. Furin processing of ADAMs mostly occurs within the secretory pathway, but some ADAMTS proteinases, such as ADAMTS5, ADAMTS7, and ADAMTS9, may be cleaved at the cell surface or extracellularly (Koo and Apte 2009; Longpré et al. 2009). Indeed, activation of stored zymogen in the ECM rather than de novo production of ADAMTS5 has been suggested as a potential mechanism for cartilage destruction in arthritis (Malfait et al. 2008; Wylie et al. 2012).
Of the four TIMPs, only TIMP3 is an ADAMTS/ADAM inhibitor; although all four TIMPs have inhibitory activity towards MMPs, certain TIMP-protease pairings (e.g., TIMP2-MMP2) show higher affinity. TIMP3 inhibitory activity towards ADAM17 and ADAMTS4 and ADAMTS5 implicates it as a key player in regulating inflammation and cartilage matrix breakdown (Kashiwagi et al. 2001; Sahebjam et al. 2007). As endogenous inhibitors of the metalloproteinases, TIMPs likely play an important role in ECM turnover of the intervertebral disc, although this has yet to be specifically investigated. The proteinase inhibitor α2-macroglobulin, which unlike TIMPs has an extremely broad spectrum of inhibition, is also capable of blocking ADAMTS activity (Somerville et al. 2004; Tortorella et al. 2004) and presumably does so in the circulation.
Despite their brief history, functional contexts were rapidly established for several ADAMTS family members, aided by their clustering into functional family groups (Fig. 8.2), stringently established associations with hereditary and acquired diseases, and via several natural and engineered animal mutations. These are summarized in Table 8.1 and described in more detail in a previous review (Apte 2009). Since some of these molecular functions clearly have potential or established relevance to the intervertebral disc, they are discussed here and illustrated in Fig. 8.3. ADAMTS1 cleaves aggrecan, versican, thrombospondin-1 and thrombospondin-2, and the cell-surface proteoglycan syndecan-4 (Sandy et al. 2001; Lee et al. 2006; Rodríguez-Manzaneque et al. 2009). It is associated with inflammation, cancer cachexia, infertility, urinary tract anomalies, and bone metastasis and has potent antiangiogenic activity (Luque et al. 2003; Mittaz et al. 2004; Apte 2009; Lu et al. 2009). Thus, ADAMTS1 could be involved not only in matrix proteolysis but also in inflammatory disc disease. ADAMTS2, ADAMTS3, and ADAMTS14 are procollagen-processing enzymes involved in excision of the amino (N)-terminal propeptide of procollagens I, II, and III (Colige et al. 1999, 2002; Fernandes et al. 2001) but could also have additional properties unrelated to procollagens. Removal of the bulky procollagen N-propeptides is a prerequisite for proper collagen assembly. In the absence of ADAMTS2, an animal disorder named dermatosparaxis results, in which skin and other collagen-rich tissues show abnormal collagen fibrils (Lapière and Nusgens 1993), although the intervertebral disc was not one of the tissues specifically investigated. Dermatosparactic collagen forms branched and thin fibrils assuming a hieroglyphic or “cauliflower-like” pattern in electron microscopy, rather than highly ordered, broad, unbranched normal fibrils. These anomalous fibrils are structurally weak, a problem most strikingly evident in the exceedingly fragile skin of dermatosparactic cattle. A corresponding human-inherited connective tissue disorder, Ehlers-Danlos syndrome, dermatosparactic type, is similar in its clinical presentation (Colige et al. 1999). However, neither spine nor disc was specifically reported to be anomalous. In collagen II-rich tissues such as cartilage, ADAMTS3 is likely to have a more significant role than ADAMTS2 because of its expression in cartilage and demonstrated ability to process procollagen II (Le Goff et al. 2006).

Table 8.1
Functions of selected ADAMTS proteins and potential relevance to intervertebral disc biology
Known functions | Potential function in IVD | |
|---|---|---|
ADAMTS1 | Cleaves versican, thrombospondin-1 and thrombospondin-2, and syndecan-4; inhibits angiogenesis. Has a role in TGFβ activation. Null mice have impaired fertility, abnormal cardiac development, and hydronephrosis | Potentially involved in ECM turnover, TGFβ activation, and angiogenesis |
ADAMTS2, ADAMTS3, ADAMTS14 | Removal of amino-propeptide of procollagens I, II, and III. ADAMTS2 mutations lead to dermatosparaxis in animals and EDS dermatosparactic type in humans | Potential role in procollagen I and procollagen II processing, collagen assembly, and maintaining tensile strength of annulus fibrosus |
ADAMTS4 | Cleaves aggrecan and versican. Null mice are reported to be developmentally normal. Combinatorial null mice (with ADAMTS1) have a thin renal medulla | Potentially involved in proteoglycan turnover in nucleus pulposus, end plate, and perichondrium |
ADAMTS5 | Cleaves aggrecan, versican, and biglycan. ADAMTS5 null mice are resistant to induced cartilage degeneration. ADAMTS5 null mice lack embryonic sculpting of pulmonic valve leaflets and have reduced interdigital web regression. Cooperates with ADAMTS9 and ADAMTS20 in interdigital web regression | Potentially involved in proteoglycan turnover in nucleus pulposus, end plate, and perichondrium |
ADAMTS7, ADAMTS12 | Reported to bind to and cleave cartilage oligomeric protein (COMP, thrombospondin-5) and granulin-epithelin precursor. ADAMTS12 null mice are developmentally normal | Potentially involved in ECM turnover in annulus fibrosus |
ADAMTS9 | Cleaves aggrecan and versican. ADAMTS9 null mice die early during embryogenesis. ADAMTS9 haploinsufficient mice have cardiovascular defects. Cooperates with ADAMTS5 and ADAMTS20 in interdigital web regression and with ADAMTS20 in closure of the secondary palate in mice. ADAMTS9 is antiangiogenic | Potentially involved in proteoglycan turnover in nucleus pulposus, end plate, and perichondrium and regulation of angiogenesis |
ADAMTS10 | Binds to and possibly cleaves fibrillin-1. Promotes fibrillin microfibril assembly. ADAMTS10 is mutated in Weill-Marchesani syndrome in humans | Potential role in fibrillin microfibril assembly |
ADAMTS13 | ADAMTS13 is required for maturation of ultra-large forms of von Willebrand factor. ADAMTS13 mutations or autoantibodies lead to thrombotic thrombocytopenic purpura | |
ADAMTS17 | ADAMTS17 mutations lead to a Weill-Marchesani-like syndrome in humans and recessive isolated ectopia lentis in dogs | Potentially involved in fibrillin microfibril assembly |
ADAMTS20 | Cleaves versican. ADAMTS20 mutations in mice lead to a white spotting mutant named belted (bt) | Potentially involved in versican turnover |
ADAMTSL2 | ADAMTSL2 binds to fibrillin-1 and latent TGFβ-binding protein 1. ADAMTSL2 mutations lead to geleophysic dysplasia. In dogs, ADAMTSL2 mutations lead to Musladin-Lueke syndrome | Expressed in IVD. Potentially involved in regulating TGFβ binding and/or activation |
ADAMTSL4 | ADAMTSL4 binds to fibrillin-1 and enhances microfibril biogenesis in cultured fibroblasts. ADAMTSL4 mutations lead to recessive isolated ectopia lentis in the eye | Potentially involved in microfibril assembly |
ADAMTSL6 | ADAMTSL6 binds to fibrillin-1 and enhances microfibril biogenesis in cultured fibroblasts and transgenic mice when it is overexpressed | Potentially involved in microfibril assembly |

Fig. 8.3
Potential roles of ADAMTS proteinases in various processes within the context of intervertebral disc components. The nucleus pulposus (NP), annulus fibrosus (AF), cartilaginous end plate (EP), and vertebral bone (B) are indicated
ADAMTS1, ADAMTS4, ADAMTS5, ADAMTS9, and ADAMTS20 have the ability to cleave the large aggregating chondroitin sulfate proteoglycans aggrecan and versican at specific sites in their core proteins (Apte 2009). ADAMTS9 is also an antiangiogenic protease that is widely distributed in microvascular endothelial cells of most organs as an apparently constitutive product (Koo et al. 2010). ADAMTS4 and ADAMTS5 seem to be the most important aggrecan-degrading proteinases (aggrecanases) in the articular cartilage degradation in arthritis (Fosang and Little 2008) and have thus been most extensively investigated in the intervertebral disc, although much on the published analysis is of gene expression and protein distribution rather than de novo functional analysis. ADAMTS7 and ADAMTS12 are unique proteinases in also being chondroitin sulfate proteoglycans (Somerville et al. 2004), the only known proteinases in mammalian genomes to have this property. They cleave cartilage oligomeric matrix protein (COMP) (Liu et al. 2006), which is an important component of cartilage ECM, as well as granulin-epithelin precursor (GEP) (Guo et al. 2010), a growth factor reported to be involved in tissue regeneration and inflammation (Bai et al. 2009). GEP was reported to act as a competitive inhibitor of ADAMTS7 and ADAMTS12 (Guo et al. 2010). Based on being a target of PTHrP and cleavage of GEP, ADAMTS7 was proposed as a negative regulator of endochondral bone formation, but this remains to be substantiated in genetic models (Bai et al. 2009; Liu 2009).
ADAMTS10 is mutated in a connective tissue disorder named recessive Weill-Marchesani syndrome (WMS). The identification of dominantly inherited fibrillin-1 mutations in WMS (Faivre et al. 2003; Sengle et al. 2012) suggested a functional link between ADAMTS10 and fibrillin-1. Such fibrillin-1 mutations typically lead to Marfan syndrome, yet WMS appears to constitute an “opposite of Marfan syndrome,” featuring short stature, short digits, stiff skin and joints, and in the eye, lens dislocation, and glaucoma. Recent work showed that ADAMTS10 bound to fibrillin-1 and fibrillin-2 was associated with fibrillin microfibrils in tissues and enhanced fibrillin-1 microfibrils formation in vitro (Kutz et al. 2011).
A unique aspect of ADAMTS proteases is the existence of a closely related family of seven independent gene products resembling ADAMTS ancillary domains (Apte 2009). These molecules, named ADAMTS-like proteins (ADAMTSL), lack catalytic domains and are therefore not proteinases; however, together with ADAMTS proteinases, they are thought to comprise a protein superfamily. Intriguingly, like ADAMTS10, the majority of ADAMTSLs, specifically, ADAMTSL2, ADAMTSL3, ADAMTSL4, and ADAMTSL6, also bind to or influence microfibril formation (Le Goff et al. 2011; Saito et al. 2011; Gabriel et al. 2012; Sengle et al. 2012) (Table 8.1). Human genetic disorders resulting from ADAMTSL2 or ADAMTSL4 mutations, i.e., geleophysic dysplasia (GD) and isolated ectopia lentis, respectively, are also caused by fibrillin-1 mutations, and both these proteins bind to fibrillin-1 (Hubmacher and Apte 2011; Le Goff et al. 2011). The mutations in GD lead to a short-stature, short-digit phenotype superficially resembling WMS (these conditions fall within a category named acromelic dysplasias), although GD is much more severe and frequently lethal in children because of cardiac involvement; however, unlike WMS, it lacks ocular involvement (Le Goff et al. 2008). ADAMTSL2 mutations in dogs also cause dwarfism and severe skin and joint stiffness, a connective tissue disorder named Musladin-Lueke syndrome (Bader et al. 2010). Analysis of cells from GD patients has suggested that there is profound TGFβ dysregulation, likely related to the pivotal role of fibrillin microfibrils in regulating TGFβ and bone morphogenetic proteins (Le Goff et al. 2008). Indeed, ADAMTSL2 also binds to latent TGFβ-binding protein-1 (LTBP1), which strongly supports a role in TGFβ sequestration in ECM or activation. We recently reviewed the strong functional involvement of ADAMTS proteinases in connective tissue regulation vis-à-vis fibrillins, specifically, in regulating the cellular microenvironment (Hubmacher and Apte 2011). ADAMTSL2 is expressed in the nucleus pulposus of the intervertebral disc (Koo et al. 2007; Sohn et al. 2010), and it could have a role in disc development because of the established role of TGFβ signaling in this process (Sohn et al. 2010).
Another cluster of ADAMTS proteinases highly relevant to IVD is the so-called proteoglycanase cluster, containing ADAMTS1, ADAMTS4, ADAMTS5, ADAMTS9, and ADAMTS20 (Apte 2004; Huxley-Jones et al. 2005). The major proteoglycan substrates of these proteinases are aggrecan, a cartilage-specific chondroitin sulfate proteoglycan, and its widely distributed relative in non-cartilaginous tissues, versican. The role of these proteinases in aggrecan destruction in osteoarthritis has been investigated by stringent analysis of null mice, biochemical assays, and association of mRNA, protein, and catabolic fragments with arthritic cartilage (Fosang and Little 2008). ADAMTS5-deficient mice are protected against either a mechanical instability-induced or cytokine-induced cartilage breakdown (Glasson et al. 2005; Stanton et al. 2005).
Although these five enzymes are potent aggrecanases, which of them has a role in physiological aggrecan breakdown, such as in skeletal development, is unclear. However, some of them were discovered to have a crucial role in turnover of versican in several developmental contexts, specifically, myocardial compaction, closure of the secondary palate, sculpting of heart valves, and resorption of interdigital webs (Stankunas et al. 2008; McCulloch et al. 2009; Enomoto et al. 2010; Dupuis et al. 2011). Two intriguing findings emerged from these discoveries: One, that proteoglycanases cooperated in versican proteolysis in the context of palate closure and web regression, and two, that a product of versican proteolysis was a matrikine with context-dependent function in cell proliferation or apoptosis in these processes (McCulloch et al. 2009; Enomoto et al. 2010). Further study of these families will no doubt find other important functions of these proteinases in normal development and disease pathogenesis.
In contrast to these functions of ADAMTS proteinases which are directly relevant to extracellular matrix maturation, assembly, and turnover, ADAMs are primarily involved in ectodomain shedding of cell-surface molecules. In particular they are identified to have a pivotal role in growth control through epidermal growth factor receptor signaling pathways, in cell adhesion, in Notch signaling, and in inflammation through processing of pro-TNF-α and cytokine receptors (Klein and Bischoff 2011; Saftig and Reiss 2011). Thus, ADAM proteinases may play a role in disc development and degeneration through modulation of these and several other pathways. Unfortunately direct effects of ADAM proteinases in the intervertebral disc have not been elucidated to any significant extent, and it remains an area of opportunity for future research.
8.3 ADAMTS Proteinases in Intervertebral Disc Biology and Disease
Three anatomic components of the intervertebral disc are relevant to its physiology and pathology in regard to metalloproteinases, namely the cartilaginous end plates, annulus fibrosus, and nucleus pulposus, which act together to fulfill the biological and mechanical functions of the disc but also differ in their matrix structure because each is mechanically specialized. The two major components of the ECM in the disc are the proteoglycan aggrecan and collagen, which are cleaved and modulated by ADAMTS proteinases. Collagen I, II, III, V, VI, IX, XI, XII, and XIV can be found in the disc at varying levels and will also change with age (Eyre et al. 2002). Of the collagens, I and II are most abundant, with I comprising the outer layers of the annulus fibrosus and II mainly located in the nucleus. Besides aggrecan in the nucleus pulposus, other proteoglycans found in lower amounts include versican, decorin, biglycan, fibromodulin, lumican, and perlecan (Roughley 2004). Aggrecan is found in both the nucleus pulposus and annulus fibrosus, mostly as aggregates complexed with hyaluronan and link protein, although it makes up a larger proportion of the nucleus pulposus (65 % dry weight) than the annulus fibrosus (15–20 % dry weight) (Le Maitre et al. 2007). The CEP of the vertebral bodies are similar in content and structure to articular cartilage found in other joints and are primarily comprised of collagen II fibers with aggrecan aggregates complexed to hyaluronan and link protein. Previous chapters in this book (Chaps. 4 and 5) provide further details regarding the roles of the proteoglycans and collagens in the intervertebral disc.
As the intervertebral disc ages, its ECM undergoes significant catabolic changes in which ADAMTS proteinases are likely to have a role. The majority of experimental studies to date have focused on the levels and localization of ADAMTS4, ADAMTS5, and catabolic products of aggrecan. Using immunohistochemistry, ADAMTS4 was detected in nondegrading disc cells from the nucleus pulposus and inner annulus fibrosus, with little immunoreactivity in the outer annulus fibrosus, suggesting a maintenance role, but in degenerated discs, the authors found an increase in ADAMTS4 that correlated with the severity of degeneration and increased TIMP1 and TIMP2 but not TIMP3 (Le Maitre et al. 2004). It was also found that the presence of inflammatory modulators such as TNF-α and IL-1 increased ADAMTS4 and ADAMTS5 expression and ADAMTS aggrecan degradation (Le Maitre et al. 2005; Séguin et al. 2005). Toll-like receptor adaptor signaling molecule MyD88 antagonized LPS or IL-1-mediated induction of ADAMTS4 and ADAMTS5 (Ellman et al. 2012). An immunohistochemical analysis of surgically resected discs showed ADAMTS4 primarily in CD68-positive mononuclear cells (monocyte/macrophages) in granulation tissue and adjacent disc, with a higher number of stained cells associated with specific herniation patterns (transligamentous extrusion and sequestration). A recent detailed analysis investigated the role of syndecan-4 in ADAMTS5 activity in nucleus pulposus cells. TNF-α and IL-1α increased ADAMTS4 and ADAMTS5 expression and promoted syndecan-4 interaction with ADAMTS5 (Wang et al. 2011). This was consistent with an association of aggrecan degradation with increased syndecan-4 and ADAMTS5 in the human intervertebral disc (Wang et al. 2011).
Stay updated, free articles. Join our Telegram channel

Full access? Get Clinical Tree








