CHAPTER 13 Laboratory tests
Introduction
• enable a definitive diagnosis to be made in situations where there may be several possible diagnoses
• measure the disease process in relation to normal parameters
• reveal occult disease processes that might affect therapeutic and treatment options
Microbiology
Indications for microbiology
• Never without good reason dismiss a microbe as a contaminant because it is not an ‘accepted’ pathogen.
• Never without good reason accept a microbe as the necessary cause of a disease merely because it is an ‘accepted’ pathogen.
Sampling techniques
Specimen types
Ideally, when microbiological information is needed, an appropriate specimen is taken from the correct site. The specimen is transported immediately to the laboratory where it is processed quickly using the best tests, which are then correctly reported; these results are returned to the originator where they can be properly interpreted at a time when the information is relevant. Thus, it is important that the results of the work done in the laboratory are a true reflection of that specimen. Reasons for failure to report an organism originally present in a specimen are shown in Box 13.1.
Box 13.1 Reasons for failure to report an organism originally present in a specimen
• The amount of specimen was insufficient
• The amount of medium into which the specimen was inoculated was insufficient
• The medium used for inoculation was of doubtful quality or unsuitable for the growth of the organism present
• The incubation time of the inoculated medium was too short or the wrong conditions were provided
• Too few colonies were examined or the organism was not recognised
It is imperative to use reputable laboratories that employ strict internal standards and where reagents and media are performance-tested using standard organisms. If this procedure is adhered to, failure to report pathogens often rests with the sampling technique employed by the requesting practitioner. It is essential to follow certain guidelines (Box 13.2) in order to obtain quality samples.
Box 13.2 Guidelines to obtain good-quality specimens
• The sample should be taken from the actual site of an infection or from where it is suspected
• Skin should not be cleaned with an antiseptic prior to taking the sample
• Strict aseptic technique must be followed to reduce the risk of the sample becoming contaminated by the microbiological flora of either the patient or the person taking the sample
• Many pathogenic organisms are surprisingly delicate. Unless special measures are taken they do not survive for long away from the body. This means it is often vital that specimens are transported to the laboratory without delay
• If some delay in transporting specimens is anticipated, it is important that steps are taken to prevent significant growth of contaminating organisms that can grow at room temperature and swamp the genuine pathogen. Suppression is normally achieved by refrigeration or inclusion of an inhibitor in the transport medium.
• It is important that sufficient sample is supplied so that the laboratory may use different methods for culture analysis of the sample and thus maximise the chances of providing meaningful results
• If at all possible, samples should be taken prior to the commencement of antibiotic therapy. A drug may suppress a pathogen sufficiently to thwart isolation and identification, without actually working well enough to allow the patient to recover
• It is often desirable for practitioners to wait for the initial results from the laboratory before starting antibiotic therapy. The results allow practitioners to choose a narrow-spectrum drug that they can be confident will do the job. However, if a life-threatening infection is suspected, then a broad-spectrum antibiotic should be prescribed without delay
• Specimens taken for microbiological analysis are by their very nature likely to contain pathogenic organisms and should, therefore, be treated with care
• Good documentation is vital to ensure that samples are not mixed up, lost or subject to inappropriate tests
• It is vital that there is dialogue between the practitioner taking the sample and the laboratory staff. For more unusual organisms the microbiologist may be able to provide advice about the most appropriate methods of sampling and transportation.
Even if the guidelines given in Box 13.2 have been adhered to, in order to be effective the laboratory requires good clinical information about the patient. It is imperative that the site of the suspected infection is stated. The symptoms should be included on the clinical history section and it is important to note recent treatment with antibiotics. Is there anything in the patient’s history or in the clinical features (e.g. colour of pus, cellulitis) that may provide a clue as to the type of organism that is causing the problem? Without this sort of detailed information, valuable time and resources may be wasted in inappropriate analyses. There is provision for all these data on the laboratory request form, which also has an integral bag for the inclusion of the specimen (Fig. 13.1).
Apparatus for obtaining specimens
Swabs
Bacteriological swabs are often made from a pledget of Dacron (Terylene) attached to the end of a holder made from wood, plastic or metal, the whole of which is sterilised before use. Cotton wool pledgets can be used, but they may release lipoproteins, which can harm some fastidious bacteria. Swabs are the instrument of choice for collection of samples where microbial contamination or infection is suspected. Sterilised swabs with or without transport media are available commercially (Fig. 13.2).
Collection of samples
Wounds and mucosal surfaces
If a large quantity of pus is present this may be drained and sent to the laboratory; otherwise, a swab should be taken. Great care should be taken to avoid contamination with the normal flora from surrounding healthy tissue. It is important that a sample is taken from the base of the wound (Fig. 13.3). If the swab is taken from the superficial edges, the flora of the adjacent skin could contaminate it. Swabs are placed into tubes containing a semi-solid transport medium, which prevents them from drying out.
Skin
For mycological (fungal) investigations, nail clippings or skin scrapings from the edge of the lesion, taken with a blunt scalpel, can be placed in either a purpose-designed packet which has a black/dark blue inner surface to help identify the sample (Fig. 13.4), or a clean, dry plastic container.
Once the sample has been obtained it should be sent directly to the laboratory. Accompanying the sample will be a laboratory request form. Accurate information will enable the laboratory staff to carry out the most appropriate tests and investigations quickly, and thus provide the clinician with the results without delay. Laboratory request forms vary, but it is important that the following information is included:
• The ward name or place where the sample was taken.
• The patient’s date of birth – resident flora change with age and it is therefore helpful to the laboratory staff in their investigations.
• Date of admission to hospital – useful for infection control staff to monitor nosocomial infection.
• Site of infection – be as specific as possible, it will help laboratory staff distinguish commensals from pathogenic flora.
• Antibiotic therapy – even small amounts of antibiotic inhibit the growth of micro-organisms in the laboratory.
• Date and time of collection of the specimen – microorganisms survive or multiply at varying rates and, thus, this information is important when the sample is cultured.
• Specimen type and investigation requested – remember most swabs look the same when they arrive in the laboratory, therefore state clearly what the specimen is. State whether the sample is for microculture and sensitivity (MC&S), for virology or serology.
• Biohazard status – if suspected, then the sample should not only be marked as such but should be transported in double-wrapped specimen bags.
Laboratory examination
Microscopy
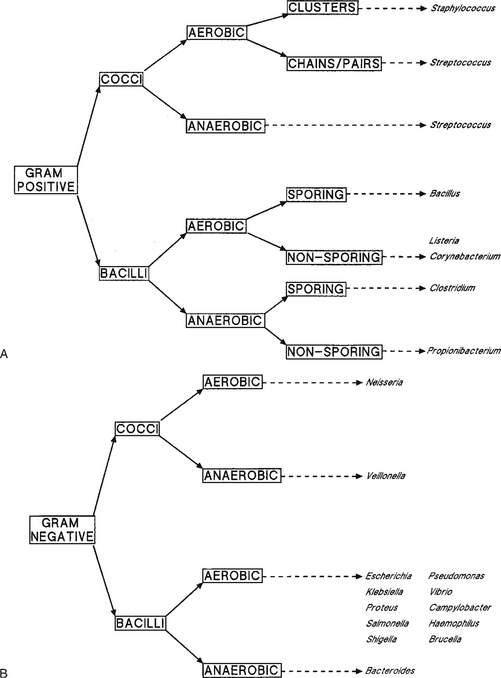
The most widely used stain is Gram’s stain: gentian violet in Gram’s stain binds to the cell wall of Gram-positive organisms and resists decolorisation with methanol or acetone. Those cells decolorised and stained with a counterstain, to make them visible, are classified as Gram-negative bacteria. The Gram stain immediately separates most bacteria into two groups and this together with other factors significantly aids diagnosis (Fig. 13.5). The other commonly used stain is the Ziehl–Neelsen stain for acid-fast bacteria such as Mycobacterium spp.
Fungal hyphae and spores can often be seen under the light microscope: skin scrapings with suspected dermatophyte infection are mounted on a slide, cleared with 10% potassium hydroxide and stained with lactophenol blue. This simple procedure can often be done by the clinician without recourse to the laboratory, thus giving an instant diagnosis. Culture of these specimens can take 2–3 weeks. Many patients will have been prescribed a topical antifungal medicament based on the clinical features of the disease which, if caused by a dermatophyte, should be well on the way to resolution by the time the results are reported.
Culture
Culture allows either the amplification of organisms initially present only in small numbers or selection of organisms from mixed inocula. Media for cultivation of bacteria and fungi must be capable of satisfying all their nutritional requirements and provide appropriate conditions which satisfy any factors which may affect their metabolism such as temperature, pH, osmolarity, oxygen, carbon dioxide and radiation.
Culture media fall into the following classes:
• Synthetic or defined media – prepared entirely from organic or inorganic chemicals such that the exact composition is known and is repeatable on any occasion.
• Routine media – prepared from a mixture of digested or extracted animal or plant protein, such as beef or soya bean. They are usually supplemented with accessory growth factors and the pH adjusted to 7.4.
• Enrichment media – usually routine media with specific additions such as whole blood, serum or additional sugars, for the growth of more exacting organisms.
• Selective media – routine media to which has been added selectively inhibitory chemicals that suppress or kill all but a few types of (known) organisms.
• Indicator media – routine media with the addition of substances which change the appearance of the media when a particular organism grows on it: for example, haemolysis of red blood cells by haemolytic bacteria.
Identification of microorganisms
Bacteria
It is first necessary to isolate the microorganism in pure culture before carrying out identification tests. This is usually achieved by streaking or spreading the initial inoculum on the surface of solid selective media to produce isolated colonies of the desired organism. A colony is then selected and may need subculturing in routine or enriched media to restore normal growth before examination. Table 13.1 lists the bacterial pathogens typically found in the lower limb. The process of identification usually involves the following steps.
Cultural characteristics
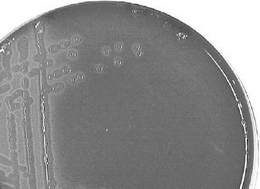
The size, shape and colour of colonies on solid media are sometimes helpful in diagnosis but are not sufficiently stable enough to be of routine value. However, the ability of an organism to grow on different media, including the stimulatory effects of added substances such as glucose, whole blood or serum, can be significant, e.g. the degree of haemolysis of blood incorporated into the media is used to differentiate streptococci. Alpha-haemolytic streptococci such as Streptococcus pneumoniae produce colonies which are surrounded by a green ring, whereas Strep. pyogenes, a beta-haemolytic bacterium that is responsible for human throat infections, grows with a clear ring around it where the blood cells have been completely lysed (Figs 13.6, 13.7). These characteristics of colony growth of bacteria are accompanied by observations of the optimum temperature and pH ranges for growth and any pigment that may be produced. The gaseous requirements of organisms can also be diagnostic. Some bacteria are obligate aerobes (e.g. Bordetella), but others (e.g. Pseudomonas aeruginosa), which usually use molecular oxygen, are capable of using nitrate if cultured anaerobically. Anaerobes are either facultative (i.e. they can grow either aerobically or anaerobically) or obligate. Other factors such as the ability to grow in the presence of antibiotics, bile salts and high salt concentration should also be noted.
Biochemical reactions
The ability of organisms to use particular substrates with a detectable end product, such as sugars, is a widely tested function. A range of these biochemical tests are available, including fermentation patterns, catalase, oxidase and nitrate production, and are, perhaps, the most useful aids to identification. For example, if a positive identification of Gram-positive cocci has been made, a catalase test can be done to aid further diagnosis. Staphylococci and streptococci are both commonly found cocci. Staphylococci are catalase-positive and are able to produce bubbles of oxygen when incubated with hydrogen peroxide. Streptococci, which are catalase-negative, do not react in this way and no oxygen is produced. This may help identify, say, a colony of staphylococci, but it does not help with identification of the species within the genus. A mannitol fermentation test may be able to determine if the colony consists of Staphylococcus aureus or Staph. epidermidis, since Staph. aureus tests positive, whereas Staph. epidermidis does not. Ready-made kits for multiple biochemical analyses are freely available commercially to enable complex tests to be carried out simultaneously with remarkable accuracy. All help to build a pattern of the metabolic activity of the organism, which – together with the information gained from microscopic and cultural examination – may be sufficient to identify the organism.
Sensitivity testing
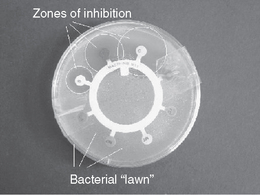
Once the organism has been identified, the susceptibility of the organism is often predictable. However, not all organisms have predictable resistance patterns and, thus, testing for sensitivity to particular antibiotics is required. Perhaps the most common method used to test antibiotic sensitivity is disc diffusion. A Petri dish is inoculated to produce a lawn of the test organism and an antibiotic-impregnated disc containing a range of antibiotics at concentrations comparable with therapeutic plasma levels is placed on the surface of the lawn (Fig. 13.8).
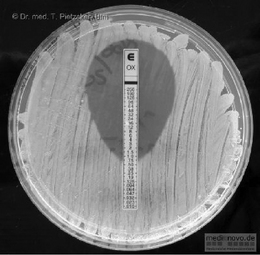
Inhibition of growth around the disc indicates the organism is sensitive to the antibiotic. Although this test indicates sensitivity of the organism, it does not show the lowest concentration (minimum inhibitory concentration, MIC) at which the antibiotic will inhibit growth of the microorganism. Although a relationship between MIC and successful outcome of antimicrobial chemotherapy cannot be clearly established, it is considered the most useful guide to the efficacy of antimicrobial therapy. Several methods of obtaining the MIC are available, and recently commercial test strips of paper with antibiotic incorporated along its length in increasing concentration have simplified the test. These test strips are put on a lawn inoculum of the test organism and the point at which the growth meets the test strip corresponds to the MIC (Fig. 13.9). From these figures the minimum bactericidal concentration (MBC) can be determined; this is defined as the lowest concentration that prevents growth after subculture to an antibiotic-free medium. These figures are required where accuracy of dose is important, e.g. in treating the immunocompromised patient.
Stay updated, free articles. Join our Telegram channel

Full access? Get Clinical Tree