First author
Year
Mouse strain
Phenotype
Outcome
Reference
Fitzgerald
2008
MRL/MpJ
Knee articular cartilage defects, ear wounds
Able to heal full-thickness knee articular cartilage defects, able to heal 2-mm ear wounds
[28]
Fitzgerald
2008
C57BL/6
Knee articular cartilage defects, ear wounds
Unable to heal full-thickness knee articular cartilage defects, failed to heal 2-mm ear wounds
[28]
Rai
2012
MRL/MpJ
Knee articular cartilage defects, ear wounds
Able to heal full-thickness knee articular cartilage defects, able to heal 2-mm ear wounds
[23]
Rai
2012
LG/J
Knee articular cartilage defects, ear wounds
Able to heal full-thickness knee articular cartilage defects, able to heal 2-mm ear wounds
[23]
Rai
2012
LGXSM-5
Knee articular cartilage defects, ear wounds
Had limited ability to heal full-thickness knee articular cartilage defects, able to heal 2-mm ear wounds up to some extent
[23]
Rai
2012
LGXSM-6
Knee articular cartilage defects, ear wounds
Able to heal full-thickness knee articular cartilage defects, able to heal 2-mm ear wounds
[23]
Rai
2012
LGXSM-33
Knee articular cartilage defects, ear wounds
Unable to heal full-thickness knee articular cartilage defects, failed to heal 2-mm ear wounds
[23]
Rai
2012
LGXSM-35
Knee articular cartilage defects, ear wounds
Had limited ability to heal full-thickness knee articular cartilage defects, able to heal 2-mm ear wounds up to some extent
[23]
Rai
2012
SM/J
Knee articular cartilage defects, ear wounds
Unable to heal full-thickness knee articular cartilage defects, failed to heal 2-mm ear wounds
[23]
Rai
2012
C57BL/6J
Knee articular cartilage defects, ear wounds
Unable to heal full-thickness knee articular cartilage defects, failed to heal 2-mm ear wounds
[23]
Rai
2012
DBA/1J
Knee articular cartilage defects, ear wounds
Unable to heal full-thickness knee articular cartilage defects, failed to heal 2-mm ear wounds
[23]
Rai
2012
DBA/2J
Knee articular cartilage defects, ear wounds
Unable to heal full-thickness knee articular cartilage defects, failed to heal 2-mm ear wounds
[23]
Eltawil
2009
C57BL/6
Knee articular cartilage defects
Unable to heal full-thickness knee articular cartilage defects
[29]
Eltawil
2009
DBA/1
Knee articular cartilage defects
Able to heal full-thickness knee articular cartilage defects
[29]
Second, in the laboratory of Francesco Dell’Accio [29], it was found that the C57BL/6 strain, like most mouse strains, does not heal articular cartilage, but the DBA/1 strain does. Longitudinal full-thickness articular cartilage injuries were generated in the patellar groove of these two mouse strains by the use of a custom-made device in which a glass bead was placed approximately 200 μm to the tip of a 26 G needle. The tip of the needle was placed anteriorly to the intercondylar notch and gently moved along the entire length of the patellar groove. The most significant findings were (1) 8-week old DBA/1 mice displayed consistent superior healing of the articular cartilage defect than C57BL/6, (2) DBA/1 mice showed a significantly less cell death and cell proliferation than C57BL/6, and (3) most importantly, the increase in articular cartilage repair was correlated with the development of OA; 8-month old DBA/1 mice failed to repair, but unlike age-matched C57BL/6, they had no signs of OA.
The above studies have used three genetically distinct mouse strains with the two strains (MRL/MpJ and DBA/1) having intrinsic repair ability compared to common C57BL/6 mouse strain. These differences in healing ability therefore provide a clue for the involvement of genetics in the ability of articular cartilage regeneration. Except for the phenotypic variation, no further insights were given for these studies from a genetic standpoint. Therefore, the need for further genetic exploration was necessary to provide a solid evidence of involvement of genetics in the response to injury. It is known that MRL/MpJ and LG/J mouse strains are closely related mouse strains since they share 75 % of their genomes identical by descent [17]. Like MRL/MpJ, LG/J mouse strain also displays complete healing of the ear wounds, including ear cartilage [31]. This raises the possibility that, similar to MRL/MpJ mice, articular cartilage may also heal in LG/J mice. To investigate this possibility, we examined the extent of cartilage regeneration in a set of common inbred mouse strains, including both healers and non-healers, and in a set of recombinant inbred lines formed from the intercross of the LG/J (healer) and SM/J (non-healer) inbred mouse strains [32]. The conceptual starting point for recombinant inbred strains is that any differential phenotype can be attributed to a restricted set of genes according to the way they have been inherited from parental strains. Therefore, we took the approach of investigating cartilage regeneration in genetic mouse models.
Third study has recently shown a variation in the magnitude and extent of cartilage healing ability among various recombinant inbred mouse strains following a full-thickness cartilage defect on the trochlear groove of distal femur. In this study, we found that some of the strains were able to heal their articular cartilage lesions superbly, others intermediately, and still others poorly. Since these mice were raised and maintained in an identical environment and subjected to same size cartilage injury (i.e., using 27 G needle) at the same age, i.e., 8 weeks, the differences in the repair response could only be attributed to the genetic diversity among the strains [23]. To this end, the broad-sense heritability estimates confirmed that variation in these mice to tissue healing is due to genetic differences among the mouse strains. There were significant differences among the inbred mouse strains in the type of cartilage formed at the site of injury (varied from typical hyaline articular cartilage to fibrous or no cartilage at all), staining intensity of proteoglycan contents (intense staining to no staining), surface regularity of injured area (smooth surface to extremely irregular), integration status of the injured area with the native health cartilage (both edges integrated to at all no integration), and finally thickness of the repair zone (in par with the native cartilage to no integration) (Fig. 13.1).
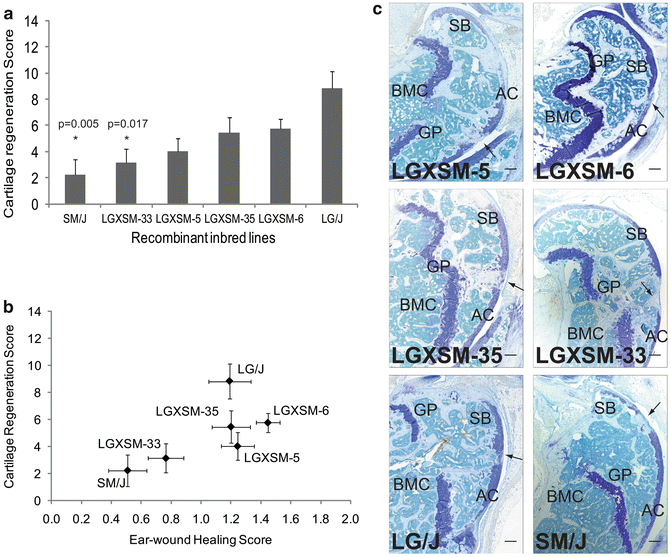

Fig. 13.1
Articular cartilage regeneration and ear-wound healing in recombinant inbred lines. (a) Articular cartilage regeneration in recombinant inbred lines showed no healing response in SM/J, LGXSM-5 and LGXSM-33 strains, while intermediate to good healing responses were observed in LGXSM-6, LGXSM-35, and LG/J strains. Asterisk with p given values indicates statistically significant difference from parental strain LG/J. (b) Correlation between knee articular cartilage regeneration and ear-wound healing showed that the two phenotypes are strongly correlated in the recombinant inbred lines. (c) Representative sagittal sections of full-thickness articular cartilage lesions from recombinant inbred lines LGXSM-5, LGXSM-6, LGXSM-33, and LGXSM-35 and parental strains LG/J and SM/J show that except for strains LGXSM-33 and SM/J, all other strains regenerated their articular cartilage and showed a deposition of proteoglycan indicating regenerative response. Arrow indicates site of defect; AC articular cartilage, SB subchondral bone, GP growth plate, BMC bone marrow cavity; bar (all panels) = 0.1 mm. Reprinted with permission from Rai et al. [23]
From these studies, it is obvious that healing capacity is dependent on mouse strains, and each mouse strain possesses a different genetic background/makeup. Having said that, it is tempting to believe that healing strength is genetically driven. The evidence presented by the above studies indicates that the ability to repair the articular cartilage is associated with the protection from OA. Therefore, the variation in cartilage injury response (i.e., the repair) and subsequent susceptibility to OA are both dependent on the genetics of the individuals.
Response of the Mouse Knee Joint to PTOA Is Genetically Modulated
So far, four studies have examined PTOA in mice from genetic standpoint. These mouse studies are described below and summarized in Table 13.2.
Table 13.2
Studies conducted to get insights into PTOA in mice
First author | Year | Mouse strain | Site | Phenotype | Outcome | Reference |
|---|---|---|---|---|---|---|
Ward | 2008 | MRL/MpJ | Knee | Articular cartilage, bone | Did not develop OA | [33] |
Ward | 2008 | C57BL/6 | Knee | Articular cartilage, bone | Developed OA | [33] |
Eltawil | 2009 | C57BL/6 | Knee | Articular cartilage | Developed OA | [29] |
Eltawil | 2009 | DBA/1 | Knee | Articular cartilage | Did not develop OA | [29] |
Hashimoto | 2012 | LGXSM-6 | Knee | Articular cartilage, bone | Did not develop OA | [35] |
Hashimoto | 2012 | LGXSM-33 | Knee | Articular cartilage, bone | Developed OA | [35] |
Lewis | 2013 | MRL/MpJ | Knee, serum | Articular cartilage, bone, synovium | Did not develop OA | [34] |
Lewis | 2013 | C57BL/6 | Knee, serum | Articular cartilage, bone, synovium | Developed OA | [34] |
First, Ward et al. [33] in the laboratory of Steve Olson investigated the correlation between intra-articular fracture healing and OA in C57BL/6 and MRL/MpJ mice. These authors experimentally created intra-articular fractures in the knees of these mice and analyzed the bone and cartilage changes by various techniques. It was found that the MRL/MpJ mouse is protected from PTOA after intra-articular fracture compared to C57BL/6. Second, Lewis and colleagues [34] again from the above research group have shown genetic and cellular evidence of decreased inflammation associated with reduced incidence of post-traumatic arthritis in MRL/MpJ mice compared to C57BL/6 mice undergoing tibial plateau fractures. These data further suggest a genetic association between joint tissue inflammations and the development and progression of PTOA in mice. Third, in the laboratory of Francesco Dell’Accio, Eltawil et al. [29] reported that C57BL/6 mice after experimental cartilage injury developed features of OA compared to DBA/1, which displayed no signs of OA. Fourth and lastly, Hashimoto et al. [35] in our laboratory used two distinct mouse strains (with the ability to heal or not to heal full-thickness cartilage lesions) to induce PTOA through destabilization of medial meniscus [36]. Changes in articular cartilage and bone showed that a mouse strain with healing ability (LGXSM-6) was protected from developing PTOA compared to the other mouse strain, which failed to heal its injured articular cartilage (LGXSM-33) and developed PTOA (Fig. 13.2). As stated above, these mouse strains are recombinant inbred strains of mice and have distinct genetic portfolio, therefore providing a direct evidence for the genetic variation in the development of PTOA in mice.
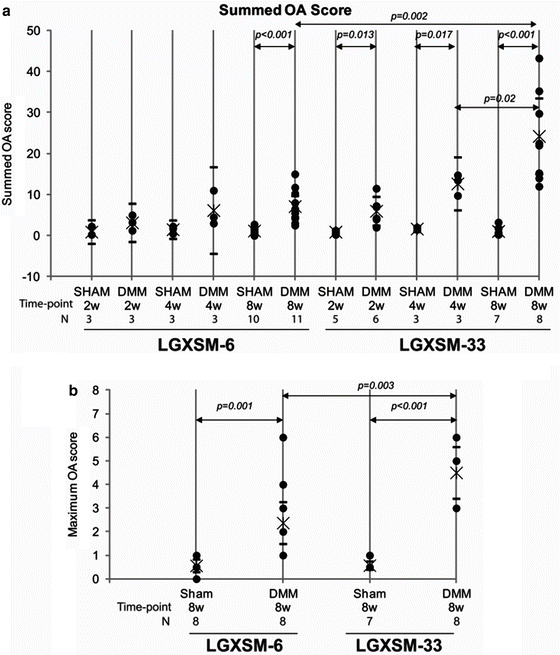

Fig. 13.2
OA score for LGXSM-6 and LGXSM-33. The sum OA score (a) from all the four quadrants and eight sections from each knee was based on cartilage damage and shows that LGXSM-33 strain overall develops more OA compared to LGXSM-6 after 8 weeks of surgery. A maximum score (b) representing highest score from medial compartment of LGXSM-6 and LGXSM-33 indicates that DMM knees had significant more OA score in both strains compared to sham knee. It also shows that LGXSM-33 has significantly higher grade of OA than LGXSM-6 at 8 weeks post-surgery. Filled circles individual data points, Star mean value, hyphens upper and lower limits of 95 % CI. Figure reprinted with permission from Hashimoto et al. [35]
Genetics of OA as Evidenced by Genome-Wide Association Scan (GWAS) Studies
Since the start of the twenty-first century, several efforts have been focused on the search for QTLs that predispose to OA. Small- and large-scale GWAS, genome-wide linkage analysis, gene-based association studies, and candidate gene studies reflect the current status quo of the genetics of complex diseases such as OA. Several GWAS studies have been conducted to understand the involvement of genetic basis of deadly human diseases. Figure 13.3 shows the number of GWAS studies for common (complex) diseases. Note that numbers of GWAS studies for OA are more than other deadly diseases such as HIV, cardiovascular problems, and obesity and lag behind Alzheimer’s disease, diabetes, and cancer.
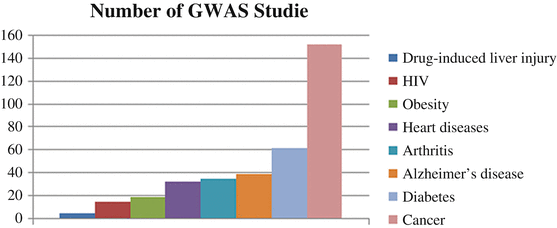

Fig. 13.3
Number of GWAS studies for the most common human diseases. These data have been taken from http://www.genome.gov and then graphed
The candidate gene association studies have been very successful since they are directed toward identifying susceptible loci for OA. While the scope of candidate gene association studies is broad, it would be injustice not to mention those studies, which have identified some of the important genes such as aspirin gene ASPN [37], SMAD3 gene [38], GDF5 gene [38–40], and FRZB gene (Table 13.3). These genes passed the strict criteria of replication in diverse Asian and European populations as well as the demonstration of functional significance. However, these findings are still debatable because of several shortcomings in the candidate gene association studies, such as lower sample size, the sparse coverage of markers employed by the relatively low-density microsatellite maps of linkage scans, and too many reports being false positive (mainly because of stringency of observed p values). These shortcomings have largely been removed by the GWAS recently (listed in Table 13.3). These GWAS studies fulfilled these criteria: genetic variants for which data was available from at least one replication cohort and P value adjusted for multiple comparisons.
Table 13.3
Candidate genes associated with OA in independent cohorts
First Author | Year | Population | SNP ID | Gene symbol | Gene name | Phenotype | P value | Reference |
|---|---|---|---|---|---|---|---|---|
Miyamoto | 2007 | Asian | rs143383 | GDF5 | Growth differentiation factor 5 | Hip OA | 1.8 × 10−13 | [41] |
Nakamura | 2007 | Asian | D14 allele | ASPN | Asporin | Knee OA | 1.0 × 10−6 | [42] |
Chapman | 2008 | European | rs143383 | GDF5 | Growth differentiation factor 5 | Knee OA | 1.0 × 10−7 | [40] |
Mototani | 2008 | Asian | rs10980705 | EDG2 | Endothelial differentiation | Knee OA | 2.6 × 10−5 | [43] |
Meulenbelt | 2008 | European | rs225014, rs12885300 | DIO2 | Deiodinase, iodothyronine, type II | Hip OA | 2.02 × 10−5 | [44] |
Miyamoto | 2008 | Asian | rs11718863, rs7639618 | VWA | Von Willebrand factor, intron A | Knee OA | 7.0 × 10−11 | [45] |
Valdes | 2008 | European | rs4140564 | PTGS2 | Prostaglandin-endoperoxide synthase 2 (prostaglandin G/H synthase and cyclooxygenase) | Knee OA | 6.9 × 10−7 | [46] |
Valdes | 2008 | European | rs4140564 | PLA2G4A | Phospholipase A2, group IVA (cytosolic, calcium dependent) | Knee OA | 6.9 × 10−7 | [46] |
Valdes | 2008 | European | rs1207421 | PARD3B | PAR-3 family cell polarity regulator beta | Knee OA | 6.0 × 10−6 | [46] |
Evangelou | 2009 | TREAT OA Consortium | rs143383 | GDF5 | Growth differentiation factor 5 | Knee OA | 9.4 × 10−7 | [47] |
Evangelou | 2009 | rs288326 | FRZB | Frizzled-related protein | Hip OA | 0.019 | ||
Valdes | 2010 | European | rs12901499 | SMAD3 | SMAD family member 3 | Knee OA Hip OA | 7.5 × 10−6 4.0 × 10−4 | [38] |
Attur | 2010 | Caucasian | rs419598, rs315952, and rs9005 | IL1RA | Interleukin 1 receptor antagonist, type I | Knee OA | <0.0001 | [48] |
Nakajima | 2010 | Asian | rs7775228, rs10947262 | HLA class II/III | Major histocompatibility complex class II | Knee OA | 2.43 × 10−8 | [49] |
Kerkhof | 2010 | Caucasian | rs3815148 | COG5 | Component of oligomeric Golgi complex 5 | Knee OA, hand OA | 8 × 10−8 | [50] |
Kerkhof | 2010 | Caucasian | rs3757713 | GPR22 | G protein-coupled receptor 22 | Knee OA, hand OA | 4 × 10−12 | [50] |
Nakajima | 2010 | Asian | rs1094726, rs7775228 | HLA class II/III | Major histocompatibility complex class II | 6.73 × 10−8 2.43 × 10−8 | [49] | |
Valdes | 2011 | European | rs143383 | GDF5 | Growth differentiation factor 5 | Knee OA | [39] | |
Kerkhof | 2011 | European | rs419598, rs315952 and rs9005 | IL1RA | Interleukin 1 receptor antagonist, type I | Knee OA | 0.006 | [51] |
Valdes | 2011 | European | rs8065080 | TRPV1 | Transient receptor potential cation channel, subfamily V, member 1 | Knee OA | 4.0 × 10−04 | [52] |
Day-Williams | 2011 | European | rs11842874 | MCF2L | MCF.2 cell line derived transforming sequence like | Knee OA | 2.1 × 10−8 | [53] |
Evangelou | 2011 | de-CODE, Rotterdam, Framingham, Twins UK | rs4730250 | PRKAR2B | Protein kinase, cAMP-dependent, regulatory, type II, beta | Knee OA | 9.2 × 10−9 | [54] |
Evangelou | 2011 | de-CODE, Rotterdam, Framingham, Twins UK | rs4730250 | HPB1 | Polybromo 1 | Knee OA | 9.2 × 10−9 | [54] |
Evangelou | 2011 | de-CODE, Rotterdam, Framingham, Twins UK | rs4730250 | COG5 | Component of oligomeric Golgi complex 5 | Knee OA | 9.2 × 10−9 | [54] |
Evangelou
Stay updated, free articles. Join our Telegram channel
Full access? Get Clinical Tree
 Get Clinical Tree app for offline access
Get Clinical Tree app for offline access

|



