3 Epidemiology and genetics of rheumatic diseases
Cases relevant to this chapter
Essential facts
1. Musculoskeletal disorders are common, with osteoarthritis affecting 80% of the population over 75 years of age and 12% of the population between 25 and 74 years of age.
2. There is a genetic component to most musculoskeletal disorders, and the genetic risk of common musculoskeletal conditions is determined by multiple genes.
3. HLA-DRB1 is the major risk gene for anti-cyclic citrullinated peptide antibody (ACPA) positive rheumatoid arthritis, but appears not be involved in ACPA-negative disease.
4. Ankylosing spondylitis has very high heritability and familiality, due to the presence of HLA-B27, but only 1–5% of HLA-B27 carriers actually develop disease.
5. Many genes have been implicated in osteoporosis, but the risk of fracture remains predominantly non-genetic.
All common rheumatic diseases have a substantial genetic component, which is not surprising given the importance of a functional musculoskeletal system in survival in times of hardship, and the inverse relationship between risk factors for autoimmunity and immunity. Genes do not, however, operate in isolation; they interact with one another and with environmental factors. Much is already known about environmental factors involved in rheumatic diseases, and the genes involved are steadily being identified.
Heritability and familiality of rheumatic diseases
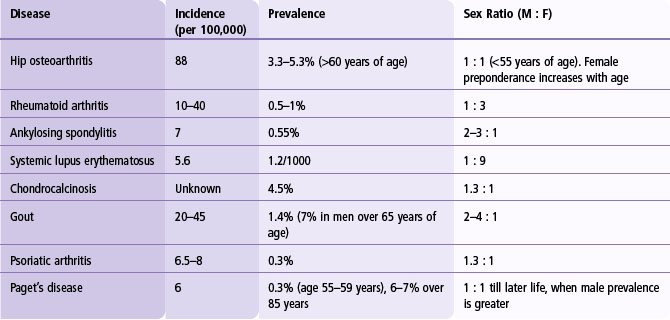
The familiality of a condition compared with the general population, termed the ‘recurrence risk ratio’, is an important statistic in determining the difficulty of mapping genes, as it is related to the heritability, number of genes involved, their population frequencies and individual magnitudes of effect. The most frequently quoted statistic to assess familiality is the sibling recurrence risk ratio, λS, which is the recurrence rate in siblings of cases, compared with the population frequency of the disease. Demographic details for common rheumatic diseases are given in Table 3.1, and heritability and sibling recurrence risk ratios are shown in Table 3.2.
Table 3.2 Heritability, familiality (assessed by the sibling recurrence risk ratio) of common rheumatic diseases
| Disease | Heritability (%) | Sibling Recurrence Risk Ratio |
|---|---|---|
| Hip osteoarthritis | ||
| Radiographic | 58 | |
| Requiring joint replacement | 27 | 1.9 |
| Osteoporosis | ||
| Twins | 80–90 | |
| Families | 40–80 | 5 |
| Rheumatoid arthritis | 60 | 5–10 |
| Systemic lupus erythematosus | 60 | 14 |
| Ankylosing spondylitis | 97 | 52–82 |
Rheumatoid arthritis
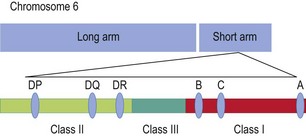
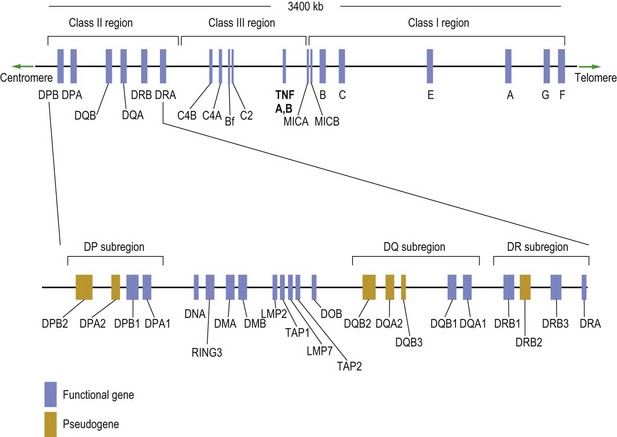
Despite case reports linking environmental exposure to infections, such as human parvovirus, Epstein–Barr virus and hepatitis B, there is no convincing evidence of a relationship between any triggering infective factor and RA. Cigarette smoking is clearly a risk factor for ACPA-positive RA, and it has been estimated that 35% of ACPA-positive RA is attributable to cigarette smoking. Genetic factors have an established role in severe disease, but genetic associations and heritability in less severe cases are much weaker. Nonetheless, strong established associations exist with MHC (major histocompatibility complex) and non-MHC genes (Figs 3.1 & 3.2 show some of the genetic sequences present on chromosome 6).
Stay updated, free articles. Join our Telegram channel

Full access? Get Clinical Tree